Characterization of a Complex Protein Mixture Using nanoLC-LIT-TOF Mass Spectrometry
Special Issues
The nanoLC LIT-TOF approach combines multiple capabilities that improve the ability to characterize complex protein mixtures significantly.
The ability to fully characterize the protein composition of biological samples holds great promise for many biochemical, biotechnical, and pharmaceutical applications. Comparison of protein expression in normal and diseased samples assists in the elucidation of disease mechanism, the determination of drug targets, and the identification of disease biomarkers. The success of these experiments relies directly on the ability to characterize, both qualitatively and quantitatively, the protein composition of biological samples. Historically, this has been an exceptionally challenging problem due to the extremely complex nature of the sample. Human plasma, for example, has been estimated conservatively to contain 500,000 protein species, not including the immunoglobulin component (1). The picture is further complicated by the extreme dynamic range of abundance. Human serum albumin (HSA) generally is present in plasma at levels of ~50 mg/mL, while tissue leakage proteins are thought to be present at concentrations of less than picograms per milliliter. This represents a dynamic range of at least 10 orders of magnitude, and the reality is that it is likely wider than even this (1). At this time, there is no technology capable of separating such a complex mixture into its individual components and detecting those components over such a large range of concentrations.
The technique that has had the most success in addressing these issues is liquid chromatography (LC) separation of the constituent peptides from a sample followed by mass spectrometry (MS) identification of those peptides. However, LC–MS is limited by both the effectiveness of the LC separation and the ability of the mass spectrometer to analyze the peptides as they are eluted from the column. Both of these areas have been the subject of intense efforts for advancement by both the academic and commercial communities. The field of LC has seen the recent advent of both nanoflow-scale chromatography (nanoLC) and ultrahigh-pressure liquid chromatography (UHPLC). MS has seen the introduction of hybrid mass analyzers that are more sensitive, accurate, and flexible than previous models, and generate data with higher information content. As instrumentation in these two fields continues to improve, we will move even closer to the goal of fully characterizing the protein composition of biological samples.
Here we describe the protein composition of an established standard protein mixture using an instrument that combines advances in both LC and MS. The system couples a nanoscale HPLC separation system with a hybrid linear ion trap time-of-flight mass spectrometer.
Instrument Overview
An overview of the nanoLC linear-ion-trap time-of-flight (LIT-TOF) MS system (NanoFrontier, Hitachi High Technologies America, San Jose, California) is shown in Figure 1. The system is comprised of the Nano LC liquid chromatograph coupled to the NanoFrontier LIT-TOF mass spectrometer. The liquid chromatograph is capable of providing steady flow rates as low as 50 nL/min while also generating a reproducible gradient. It achieves these flow rates using a splitless architecture and is designed to enable more precise and stable flow rates than systems that employ a splitter to achieve nanoscale flow.
The gradient is formed using the system's Dual Exchange Gradient System (DEGS), illustrated in Figure 1a. In this scheme, one pump (the gradient pump) forms the gradient at a high flow rate (typically 100 μL/min), while a second pump (the nano pump) is pumping constantly at a low flow rate (50–250 nL/min). The two pumps interface with one another at a 10-port valve. The valve is plumbed with two loops in such a manner that the gradient is always pumping through one of the loops while the nano pump is pumping through the second loop. The 10-port valve is switching constantly back and forth every minute. In this way, the nanopump is always pushing the gradient solution that was deposited previously in a given loop to the column, while the gradient pump is concurrently pushing its solution into the second sample loop. In this alternating fashion, the gradient is delivered to the column at nanoscale flow rates.
Figure 1b schematically illustrates the ion optics of the mass spectrometer. As ions are formed, they are funneled initially into a linear ion trap. The ion trap provides MSn capability to the system. It is also the first of three possible fragmentation mechanisms available on the instrument, utilizing He gas to promote collisionally induced dissociation (CID). For peptides, these fragments usually take the form of b and y ions, from which peptide sequence data can be reconstructed. Upon ejection from the trap, the ions then can be directed into the electron-capture dissociation (ECD) cell. ECD is a soft fragmentation method that generates c and z ions from peptides. Because it is a nonergotic fragmentation method (2), it tends to leave posttranslation modifications (PTMs) intact, making this an extremely useful technique for characterizing phosphorylation, acetylation, glycosylation, and other PTMs. After the linear ion trap and optional ECD fragmentation, ions enter the quadrupole collisionally induced dissociation (QCID) cell, in which helium is again used to fragment the analytes. Because it is a quadrupole, the QCID cell allows for the observation of lower mass fragments than those formed in the linear ion trap, resulting in more complete sequence information than possible with the linear ion trap alone. Finally, ions are guided into the time-of-flight tube for mass analysis. The time-of-flight analyzer provides high mass accuracy, exceptional resolution, and excellent sensitivity, which greatly assists in the analysis of complex mixtures.
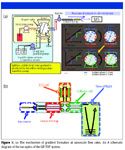
Figure 1
Experimental Description
Universal Proteomics Standard 1 (Sigma, St. Louis, Missouri) is a mixture of 48 known human proteins, present in equal concentrations. It has been shown to be an excellent sample for the diagnosis and evaluation of mass spectral instrumentation (3). Briefly, 5 pmol of the UPS-1 standard was dissolved in 50 μL of 8 M urea, and subsequently diluted in 150 μL of 50 mM ammonium bicarbonate. A 100-μL volume of the sample was reduced by the addition of 25 mL of 10 mM dithiothreitol followed by incubation at 56 °C for 1 h. Alkylation was carried out by the addition of 25 μL of 50 mM iodoacetamide followed by incubation at room temperature for 1 h in the dark. A 15-μL volume of acetonitrile was added to promote digestion. Digestion was performed by the addition of 20 μL of a 25-μg/mL solution of trypsin (Roche, sequencing grade), followed by incubation at 37 °C overnight (17 h). The digestion reaction was quenched by the addition of 2 μL of acetic acid. The resulting solution was speed-vacuumed to a final volume of 100 μL with a peptide concentration of 25 fmol/μL.
Three separate LC–MS runs were performed, in conjunction with information based acquisition (IBA). IBA is a data-dependent filtering algorithm that prevents abundant peptides from being analyzed repeatedly within the same LC–MS run as well as over a series of separate LC–MS runs, increasing the number of unique peptides analyzed by the instrument. Peptides were separated on a 150 mm × 0.05 mm MonoCap C18 column (GL Sciences, Tokyo, Japan) using a 5–40% acetonitrile gradient over 120 min. The runs contained 25, 50, and 125 fmol, respectively, of digested protein (for a total of 200 fmol). The basepeak ion chromatograms for these runs are shown in Figure 2. The resulting MS-MS data were searched using the Mascot search engine (Matrix Science, Boston, Massachusetts) against the human SwissProt database (release 53.0) using mass tolerances of 0.5 Da for both peptide and fragment mass values.

Figure 2
The LC–MS analysis found evidence of 41 of the 48 proteins present in the standard sample. Of these 41 proteins, 37 exhibited statistically significant Mascot scores. While multiple peptides were observed for the other four proteins, their overall scores fell under the threshold for positive identification. These proteins are listed in Table I, along with their total Mascot scores and sequence coverage values.

Table I
Conclusion
The nanoLC LIT-TOF approach combines multiple capabilities that improve the ability to characterize complex protein mixtures significantly. The LC system provides splitless nanoscale flow rates, improving sensitivity. The hybrid MS analyzer further increases sensitivity while concurrently improving resolution and mass accuracy. The hybrid configuration results in a flexible approach capable of many types of experiments, including simulated multiple reaction monitoring and neutral loss experiments. ECD fragmentation is a powerful technique for elucidating the structure and position of PTMs.
In combination, these capabilities result in an instrument that is exceptionally well suited for protein and peptide analysis, as shown by the analysis of Universal Protein Standard 1. Of the 48 proteins that comprise UPS-1, 37 were identified positively over the three LC–MS runs. Peptides from an additional four proteins were observed, but their Mascot scores fell under the probability threshold to be considered positive identifications. These results were generated using only 200 fmol of total protein digest.
Acknowledgment
The authors would like to thank Thu M. Doan of UCLA for her help.
References
(1) N.L. Anderson and N.G. Anderson, Mol. Cell Proteomics 1, 845–867 (2002).
(2) Stone D.-H. Shi et al., Anal. Chem. 73(1), 19–22 (2001).
(3) P.C. Andrews et al., sPRG2006 Study: A Proteomics Standard. Presented at the ABRF 2006 Conference, Long Beach, CA, February 11–14, 2006.
M. Alexander Shaw, Roselle Visaya, and Chad Ostrander are with Hitachi High-Technologies America, San Jose, California. Masaki Watanabe is with Hitachi High-Technologies, Hitachinaka, Ibaraki, 312-8504, Japan. Stone D.-H. Shi and Michael J. Greig are with Pfizer Global Research & Development, La Jolla Labs, San Diego, California.

Polysorbate Quantification and Degradation Analysis via LC and Charged Aerosol Detection
April 9th 2025Scientists from ThermoFisher Scientific published a review article in the Journal of Chromatography A that provided an overview of HPLC analysis using charged aerosol detection can help with polysorbate quantification.
Analyzing Vitamin K1 Levels in Vegetables Eaten by Warfarin Patients Using HPLC UV–vis
April 9th 2025Research conducted by the Universitas Padjadjaran (Sumedang, Indonesia) focused on the measurement of vitamin K1 in various vegetables (specifically lettuce, cabbage, napa cabbage, and spinach) that were ingested by patients using warfarin. High performance liquid chromatography (HPLC) equipped with an ultraviolet detector set at 245 nm was used as the analytical technique.
Removing Double-Stranded RNA Impurities Using Chromatography
April 8th 2025Researchers from Agency for Science, Technology and Research in Singapore recently published a review article exploring how chromatography can be used to remove double-stranded RNA impurities during mRNA therapeutics production.











