Advanced Profiling Method based on MELDI–MS For High-Throughput Pattern Analysis in Proteomics
LCGC Europe
A new proteomic profiling method based on material-enhanced laser desorption/ionization (MELDI) has been developed to identify candidate biomarkers that are selected in MELDI mass profiles. The basic principle of applying MELDI is to trace out the low-concentration species generated as a result of disease, which can then be used as diagnostic markers after their authentic validation. The first step of the MELDI approach is applied to reduce the complexity of proteomic samples by specific binding of serum proteins onto chemically modified MELDI beads, which are then directly analysed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS). The resulting mass profiles provide the basis for differentiating diseased samples from control samples. The use of liquid handling robots for sample preparation for high-throughput applications leads to higher reproducibility, which is crucial to succesfully identify disease markers. The ongoing development of MELDI for the..
Cancer can, in many cases, be cured if detected at an early, organ-confined stage. To help with early detection, there are considerable efforts to develop new potential biomarkers that improve current diagnosis and prognosis methods for different diseases. Proteomics plays a major role in obtaining further insight into fundamental biological processes and relationships for disease diagnosis.1,2 The major target application of proteomics is to evaluate disease related markers as over- or under-expressed proteins, which help to distinguish between healthy and diseased samples for early diagnosis and can even determine how far the disease has advanced. Although several biomarkers for tumour diseases, such as the prostate-specific antigen (PSA), the carcinoembryonic antigen (CEA) or the alpha-fetoprotein (AFP) have been identified and introduced successfully into clinical practice, their sensitivity and specificity have been limited.

A good example is prostate cancer, the most frequently diagnosed cancer and the second leading cause of cancer death in men in western countries.3 The prostate marker PSA is quite sensitive, however, it does not correctly differentiate benign from malignant prostate disease, and can miss some significant prostate cancers.4,5 Thus, further effort is warranted to search for additional markers to improve disease specificity. In this respect it is very likely that multiple biomarkers will be required to improve early detection, diagnosis and prognosis. This, in turn, requires sophisticated analytical strategies and workflows, including fractionation as well as pre-concentration steps to reduce the sample complexity and increase the relative content of low-abundant species. Further efficient separation as well as detection and post-experimental evaluation methods are also required.

KEY POINTS
In general, workflow strategies in proteome research can be divided into top-down and bottom-up approaches.6,7 Top-down strategies refer to all methods and proteome workflows, which use intact proteins for a first separation or fractionation step. Consequently, all gel-based, screening (protein profiling) as well as liquid chromatography (LC)-based methods of intact proteins have to be considered in this context.8,9 On the other hand, bottom-up strategies investigate proteins by analysis of their respective peptides, involving the enzymatic digestion of the whole samples to obtain smaller peptide fragments that are still sufficiently distinctive for protein identification.10,11
One of the most classical top-down techniques for discovering disease-associated proteins is two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) followed by the detection and identification of multiple protein species by MALDI-TOF-MS.12,13 This technique is unchallenged in its ability to resolve thousands of proteins but it is laborious, requires large quantities of protein, lacks critical reproducibility, lacks standards and it is not easy to convert the results into a routinely used diagnostic test.
New bioanalytical methods based on mass spectrometry could provide diagnostic and prognostic information for cancer and other disease-related biological fluids. However, without a separation process beforehand, mass systems have continued to be limited in their ability to analyse and identify potential markers. A major reason is the extremely high complexity of protein samples with a dynamic range up to 1012. Blood serum seems to be an ideal clinical sample for proteomic analysis because it can be easily collected from the patient and has a very high protein concentration (60–80 mg/mL). Nevertheless, only 22 proteins make up 99% of the whole serum proteome and many potential biomarkers are likely to be present at lower protein concentrations.14 Therefore, several sophisticated approaches such as LC-electrospray ionization (ESI) MS15 have been developed as sensitive detection tools to resolve and visualize complex peptide and protein mixtures over a broad mass range. In general, LC–MS methods generate large and highly complex data sets, which require powerful algorithms and software tools to handle and analyse them.16 Furthermore, LC–MS strategies are not yet very suitable for screening a large numbers of samples due to time-consuming analysis. However, screening by profiling methods enables the analysis of numerous samples in a short period of time and can therefore be employed for mining out differences between a huge number of deceased and healthy samples. Moreover, profiling methods allow the simultaneous measurement of a range of markers that result in, statistically, more stringent differentiation and a better classification of patient groups.17 For example, mass fingerprinting of blood samples can lead to some disease markers because the leakage of peptides and proteins into the bloodstream by cells, in response to a disease, can be used as markers.18,19
In this review, we want to provide an overview about different profiling strategies based on a new approach termed as material-enhanced laser desorption/ionization (MELDI).20 MELDI has been introduced as a mass spectrometric-based technique for pattern analysis of biological samples. While being similar to other approaches such as surface-enhanced laser desorption/ionization (SELDI) 21 or protein profiling by magnetic beads,22 MELDI enables both screening of biological samples, as well as identification of serum constituents in conjunction with LC–MS.
Sample Collection, Handling and Storage
Several studies have examined the effects of pre-analytical procedures including protein immobilization,23 matrix application24 and sample processing25 by determining their relative influence on the results. With the extra sensitivity of MS, it is likely that even quite small changes during sample collection and storage, particularly those leading to degradation and breakdown into smaller fragments, might be detected. In many cases biological fluids like serum, plasma or urine are investigated to monitor diseases at their different stages, because they might be rich sources of potential disease markers that could reflect the ongoing physiologic state of an individual organism.26 Blood serum especially, is used for many analytical approaches because it contains a total of 60–80 mg/mL of protein with a concentration range spanning up to twelve orders of magnitude. As blood perfuses tissues and organs, proteins and many other compounds, which are secreted or lost from damaged or dying cells, are released into the blood circulation after cleavage processes into smaller protein fragments.27 Many researchers believe that an effective characterization of potential disease markers will require methods to remove the most abundant proteins prior to analysis.
However, the reproducibility and effectiveness of many depletion methods have always been questioned and there are serious concerns that potentially important markers which are associated with high abundant carrier proteins (e.g., albumin) could be removed by depleting them.28 Moreover extensive sample handling during the depletion process increases the chance of sample loss, protein degradation and modification artifacts, resulting in substantial sample-to-sample variation. Pre-analytical aspects, such as centrifugation (speed, time and temperature), storage time and temperature, exposure to freeze–thaw cycles are also likely to be important, and require further investigation or, at least, consistency within studies. An interesting observation was made by Villanueva et al.,19 who provided evidence that disease markers might be generated after the patient's blood collection ex vivo as a result of proteinase-mediated enzymatic cleavage. The impact of these results in biomarker discovery is quite significant as it is widely believed that ex vivo proteolysis should be suppressed because it destroys endogenous biomarkers. Therefore the authors recommend that after blood sampling, disease-specific proteinases should not be suppressed by the addition of proteinase inhibitors as it might prevent the generation of disease markers.
The most common variations between laboratories and/or medical institutes include blood-sample tubes and other preparation aspects.29 There are studies examining whether different types of blood-collection tubes add molecules to specimens that might appear as interfering or confounding peaks during MS profiling.30 Drake et al.,31 concluded that most types of commonly used blood-collection tubes for serum collection add polymeric components that can be detected by mass spectrometry covering the m/z range of 1000–3000. These peaks potentially complicate and compromise the interpretation of profiles in the low-molecular-mass range. Very often silicones are used as lubricants for stoppers or coatings for the internal surface of tubes, and can confound peaks, as well as the polymeric surfactants such as polyvinylpyrrolidones or polyethylene glycols, which might be added to influence surface wetting.
Surface-enhanced Laser Desorption/Ionization Mass Spectrometry (SELDI)
Surface-enhanced laser desorption/ionization (SELDI), which was originally introduced by Hutchens and Yip in 1993,32 has proven to be an effective tool for purification, selective enrichment as well as pre-concentration of biological samples prior to MS evaluation. SELDI is the technology using MALDI supports with different chromatographic affinities to specifically capture and enrich biomolecules as described for the ProteinChip Arrays (Ciphergen Biosystems, Fremont, CA, USA).33 Compared with conventional MS-applications, the SELDI-technology is very easy to handle and timesaving regarding sample preparation and analysis.34 In general this method consists of a SELDI Chip, a TOF mass analyser and software for data collection and analysis.35 These chips offer a range of chemical affinities like hydrophobic, cation- and anion exchangers, IMAC, antibody-antigen and DNA-protein to immobilize specific proteins and to analyse and identify them through MS. Different mass fingerprints are generated from biofluids which can be furthermore classified as healthy or diseased by applying adequate bioinformatic tools.36
Differences in protein-profiles of disease and control-related samples are caused by overexpression, abnormally shed proteins or protein fragments, modified proteins, proteolytically cleaved proteins or degradation due to the proteosome pathway.37 There have been many examples of the use of SELDI for the determination of disease biomarkers, with the primary focus being diagnostics for many forms of cancer. During the last years SELDI technology achieved few authentic goals by claiming some potential biomarkers for Alzheimer disease,38 amyotrophic lateral sclerosis (ALS) disease,39 ovarian cancer40 and prostate cancer.41 However, the enthusiasm for using this technology declined because of methodological and bioinformatic artifacts and biases found by other groups, which questioned the validity and reproducibility of the published results.42,43
Additionally, concern has been raised about the long-term robustness of SELDI and the possible contributions of non-biological variation to the results.44,45 A major question is: Is just mining out the differences between protein-patterns of healthy and diseased serums enough to incorporate this method into clinics? By performing solely protein profiling, predictions are only based on the comparative pattern analysis of healthy and diseased samples and the question still remains, whether these distinctive peaks, picked out through SELDI-MS without their identification, are clinically applicable for diagnostics. Another important consideration is that the specificity of biomarkers must be checked by comparing them from one disease with other similar diseases. Although SELDI technology is not easily applicable to identify candidate disease markers it can be used as a first step towards biomarker discovery for fast protein screening of biological relevant samples.
Material-enhanced Laser Desorption/Ionization Mass Spectrometry (MELDI-MS)
The main focus of the emerging MELDI-MS approach is to screen biofluids like serum or urine to explore distinctive peaks among healthy and diseased samples, which can then be used as diagnostic markers after their authentic validation and identification. A major advantage of MELDI compared with ProteinChip-based SELDI-MS technology is the application of particles (MELDI carriers) and the sample preparation in suspension.46 Due to the three-dimensional shape of MELDI materials, they provide more surface area, resulting in higher peak capacity and MS sensitivity (Figure 1). Multiplexed protein pattern analysis based on material morphology, physical characteristics and chemical functionalities provide a multitude of protein patterns. MELDI comprises different materials such as modified cellulose beads, silica, organic polymer beads or carbon nanomaterials as carriers for protein binding (Table 1).
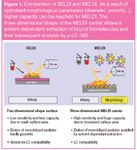
Figure 1
In a first step the MELDI approach is applied to reduce the complexity of proteomic samples by specific binding of serum proteins onto chemically modified MELDI beads, which are then directly analysed by MALDI-TOF-MS [Figure 2(a) and 2(b)]. The resulting mass profiles provide the basis for differentiating diseased from control samples. In a further step the analytical strategy includes the solvent dependent extraction of bound biomolecules [Figure 2(d)] followed by LC separation [Figure 2(e)] hyphenated to offline-MALDI [Figure 2(f)] or online-ESI MS–MS [Figure 2(g)] for their subsequent identification. In an alternative approach, the immobilized proteins can be enzymatically digested on-bead [Figure 2(c)] and subjected to MS or LC–MS analysis. Finally, advanced bioinformatics tools are used for quantification and comparison of individual proteins and up or down-regulation domains between diseased and control samples. The following sections will give a short overview of all the different MELDI carriers including their development and application:

Table 1: Overview of the described MELDI materials. Each specific application demands the most suitable carrier.
Modified cellulose beads: The ability of MELDI to distinguish prostate cancer from control was proved by a comprehensive biomarker study analysing lots of serum samples. Prostate cancer is the most common malignancy in men in western countries47 and early diagnosis is still based on the serum test for prostate-specific antigen (PSA), a test with limited disease specificity (35%).48 Therefore, the search for new and more reliable biomarkers to stratify disease onset and progression remains a challenge. It is very likely that multiple biomarkers will be required to improve early detection, diagnosis and prognosis. For a MELDI-based biomarker study on prostate cancer, spherical cellulose beads (8 μm in diameter) with IMAC-Cu2+ functionalities [Figure 3(a)] were synthesized to specifically bind serum constituents.49 Mass profiling was performed without a prior albumin or immunoglobulin depletion as potential disease markers might be associated with albumin or other high abundant serum proteins. Sample preparation was performed on a liquid handling robotic system by using special micro columns containing 1.5 mg of the cellulose resin. The micro columns have a permeable membrane, allowing the sample to easily pass through and maximizing the capture efficiency [Figure 3(b)].

Figure 2
The protein-loaded cellulose beads were directly analysed by MALDI-TOF MS. Mass spectra were processed using different statistical tests to differentiate prostate cancer from non-cancer, within a group of 137 cancer and 163 control serum samples. In a first step, data were processed by a two-sample Kolmogorov–Smirnov goodness-of-fit test (briefly, KS-test) to reduce data to pick-out useful features. In a second step, an empirical cumulative distribution of the coefficient of variation (CV), which suggests a threshold to reduce the effect of other diseases affecting some of the high-risk controls, was performed. To further compress and denoise the data discrete wavelet transformation (DWT) was used. Finally, the experiment was repeated 200 times independently, each time the classifier was trained on a 10-fold x-validation, which resulted in a clinical sensitivity and specificity of higher than 90%. Figure 3(c) shows the MALDI mass spectra of a control serum [Figure 3(a)] and a prostate cancer serum [Figure 3(b)] where the discriminating m/z signals were labelled.

Figure 3
Modified polymer beads: In this particular study, polymeric sample supports based on glycidyl methacrylate (GMA) and divinyl benzene (DVB) were synthesized and subsequently modified to get different affinities such as IMAC, reversed phase- (RP) and anion exchange- (AX) chromatography.50 Mass pattern analysis was carried out and the incubation time could be reduced to 1 min, allowing a fast screening of many samples. As a large percentage of the total number of peptides is not observed by a single MALDI MS analysis, the analytes were eluted from the polymer surface and subjected to reversed-phase μ-HPLC using a poly(styrene/divinyl benzene) (PS/DVB) monolithic column for further preconcentration and fractionation. The fractions were spotted directly on a MALDI target and MALDI matrix was added. Each fraction was subjected to acquire tandem TOF fragment ion mass spectra followed by database searching analysis using Mascot search engine for their identification.51 The combination of MELDI and LC-MALDI MS–MS by using modified poly(GMA/DVB) beads could be successfully applied for protein profiling as well as for the identification of serum peptides in the low-molecular-weight proteome (< 4000 Da).
Modified silica particles: Silica particles of different porosity were functionalized with iminodiacetic acid (IDA) and loaded with Cu2+ ions to yield Cu2+ -IDA-silica for their further application as MELDI carriers.52 In this study the effect of the material's pore size on the MELDI profiles was investigated. The pore size of the MELDI material affected the mass profiles to a great extent, showing that surface morphology is another parameter that has to be considered in addition to surface chemistry (Figure 4). Since loading capacity and chromatographic efficiency are strongly affected by the porosity of the stationary phase, differences in the mass spectra obtained by direct irradiation of the particles after serum incubation and matrix mixing could be observed. Cu2+ -IDA-silica materials with small pores in the range of 60–120 Å were shown to give poor mass spectra (low signal intensity and low number of detected masses) even though providing high surface area and high capacity. The application of wide-pore Cu2+ -IDA-silica materials (1000 Å), however, resulted in high signal intensities but suffered from number of detected signals. Ideal porosity was found to be at 300 Å, providing reasonable intensity and maximal number of detected signals. Furthermore, the evaluation of protein adsorption isotherms provided information on the binding properties towards proteins. Therefore, the binding capacity of hen egg white lysozyme (HEWL) was investigated for Cu2+ -IDA-silica using the Langmuir's adsorption theory and resulted highest capacity at a mean pore diameter of 120 Å, reaching almost 350 μg/mg.

Figure 4
Functionalized C60-fullerenes: Since their experimental discovery in 1985,53 fullerenes have attracted considerable attention in medical science.54 Investigations of chemical, physical and biological properties of fullerenes have yielded promising information and their unique carbon cage structure coupled with the immense scope for derivatization make fullerenes an interesting material for biological applications. In a special case three different C60-fullerene derivatives were used as MELDI carriers for the identification of low mass serum constituents.55 After the selective enrichment of diluted human serum on dioctadecyl methano[60]fullerene and [60]fullerenoacetic acid, protein profiling was performed by direct particle irradiation in the preferred m/z range from 2–30 kDa by MALDI-TOF MS. In a parallel approach the immobilized serum constituents were eluted from the C60-fullerenes and subjected to a μHPLC system. For high speed separation a monolithic stationary phase based on poly(p-methylstyrene-co-1,2-bis-p-(vinylphenyl)ethane (MSt/BVPE) was applied.56 The LC-eluent was then directly spotted onto a MALDI support followed by the addition of CHCA matrix for subsequent MALDI MS–MS analysis. It could be shown that MS/BVPE-based capillaries significantly increased the number of detectable mass signals in the mass range from 1–4 kDa and improved the mass resolution and the signal-to-noise (S/N) ratio by around 140–200%.
Functionalized graphitic nanofibres: Graphitic nanofibres (GNFs), 100–200 nm in diameter and 5–20 μm in length were modified in order to yield different affinities (Cu2+ and Fe3+ loaded IMAC as well as cation and anion exchange materials) for the immobilization of a range of biomolecules from biofluids.57 GNF exhibits cylindrical and conical structures, being ascribed by stacked curved graphite layers that form cones or cups.58 The sp2 carbon units are geometrically arranged into symmetries and provide a number of reactive sites for further derivatization processes. This exceptional structural constitution causes GNF carbon materials to possess extraordinary electrical, mechanical and thermal properties, which might advantageously influence the adsorption of organic compounds onto the carrier material59 as well as the desorption/ionization process in MALDI-TOF MS by improving the energy-transfer efficiency.60 The analysis of a series of standards and serum samples by using the MELDI approach clearly demonstrated that derivatized GNFs exhibit appropriate qualities to immobilize a selective range of proteins and peptides through their special affinities and natural characteristics. Remarkable peptide and protein-loading capacities were quantified through HPLC studies resulting in a capacity of 30 μg insulin per mg GNF.
Functionalized carbon nanotubes: The discovery of carbon nanotubes as "multi-walled carbon nanotubes (MWNT)" by Lijima in 199161 and "single walled carbon nanotubes (SWNT)" independently by Lijima and Bethune et al. in 199362 has given a new biologically important material to be used in bioanalytical research. As the name implies, the SWNTs are made up of single cylindrical graphitic layers whereas MWNTs consists of multiple ones. Carbon nanotubes (CNT) are the hollow cylindrical form of carbon, rolled in the form of tubes of hexagonal carbon rings, whereas the ends are composed of hexagonal rings covered by pentagonal rings. For their application as MELDI carriers, purified carbon nanotubes (CNTs) were oxidized with acidic mixture under intense harsh conditions, acid chlorified, derivatized with IDA and loaded with Cu2+ ions.63 The Cu2+ -IDA immobilized CNTs were treated with human serum and directly analysed from 2–10 kDa by MALDI-TOF MS. The most exciting feature of derivatized nanotubes is their excellent efficiency to work in the higher mass range. The most exciting feature of derivatized nanotubes is their excellent efficiency to work in a higher mass range. Human serum albumin (66 kDa) especially, could be perfectly detected by employing the described nanotube derivative.
Derivatized diamond: Diamond is an exceptional material that shows interesting properties such as high stiffness, thermal conductivity, optical transparency range, physicochemical stability and erosion resistance and inertness.64 For the application of diamond as a MELDI carrier, diamond powder (< 50 nm) was first oxidized, acid chlorified, derivatized with IDA and loaded with Cu2+ ions.20 The huge surface area (200–450 m2 /g) provides a number of sites for functionalization and subsequent binding of proteins from high concentrated serum samples. However, problems are faced during sample preparation when diamond agglomerates, especially due to electrostatic forces. This is a general problem which one can face by suspending nanoparticles. The functionalized diamond powder mainly binds his-tagged proteins through coordinate bonds with loaded Cu2+ on the chelating ligand (IDA). However, the high loading capacity of derivatized diamond powder may include some proteins which are non-specifically bound, as the potential of carboxylated diamond for binding proteins is already reported.65
Automation of MELDI
Pattern analysis is negatively influenced by the biological variance of the samples and the analytical variance of the measurement. To decrease the biological variance, a well organized working plan, including critical evaluation of sample quality such as a complete history record, sampling condition, sample transportation, pre-treatment and storage, is needed. To minimize the analytical variance and to increase the robustness of the method, a complete automation of all steps is of the utmost importance. Therefore, all steps starting from sample preparation, extraction, sample spotting and MS analyses were fully automated. The MELDI sample preparation supports automated routine analysis by means of liquid handling robots for high-throughput applications leading to higher reproducibility which is crucial for a successful identification of disease markers. By using an interfaced robotic handling system, more than 100 samples can be processed in a day. For automation micro columns are first packed with MELDI resin. All steps including equilibration, incubation, washing and on-target spotting of the MELDI resin are carried out by the robotic system to achieve the highest reproducibility. Matrix is spiked with internal standards (angiotensin I and cytochrom C) to achieve high-quality mass spectra having less mass shift due to recalibration software.
Lable-free Quantification by μLC–MS
For screening proteins in biofluids, direct laser irradiation of the adsorbed proteins is possible over a mass range of m/z = 2000–20000. However, for identification, it is necessary to elute the adsorbed compounds and subject them to MS–MS analysis. In an alternative strategy the immobilized proteins get tryptically digested on the MELDI carrier, followed by μLC–MS and MS–MS analysis. This approach was found to be be highly efficient and does not require any further elution or desalting step. Separation is performed on a poly(MSt/BVPE) capillary column which provides increased separation speed due to its good hydrophobic properties, allowing the use of steep gradients and reduced equilibration times. The on-bead digested serum samples are label-free quantified and identified via μLC–MS followed by statistical data evaluation, delivering information about over- and under-expressed peptides and proteins. Therefore, proper LC–MS software is applied, creating a combined peptide map from all samples to detect regulated signals and identify the corresponding proteins by database search (Figure 5). The detection algorithm handles complex samples and overlapping peptides, while accurate quantification and normalization enables confident differential analysis. Peptide ions of interest are taken forward for multivariate statistical analysis and identification.
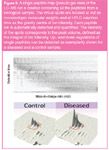
Figure 5
Conclusions
A material-based approach called material-enhanced laser desorption/ionisation mass spectrometry (MELDI-MS) was recently introduced to screen biological fluids for biomarker discovery. One of the main benefits of MELDI is the application of different carrier materials with high capacity and surface functionality to bind a multitude of serum constituents. In MELDI, the particle morphology beyond surface chemistry has been introduced for enhanced multiplexed protein profiling. During the evaluation process of various functionalized carrier materials, it was observed that not all the materials provide the same mass fingerprints. For example, cellulose carrier materials of different shapes resulted in unique peak patterns, 8–10 μm size range being the most effective in terms of number and signal intensity of resulting peaks.
As the particle size increased from 20–30 μm, or the shape changed from spherical to fibrous, the efficiency decreases tremendously, suggesting that fibrous and large spherical cellulose particles are not adequate for MELDI. Furthermore, the porosity of the investigated MELDI materials had an extreme influence on the binding properties and appearance of the resulting mass spectra. Narrow pore silica particles
(60 Å) did not provide adequate MS traces. However, particles with 30 nm pore size resulted in high-quality spectra with the maximal number of detected signals. The capacity of MELDI materials is high and the sensitivity achievements of < 1 fmol/μL can ensure the binding of low abundant components. Sample preparation protocols were further optimized and incubation times could be reduced to a few minutes as reported for poly(GMA/DVB) derivatives. A major advantage of organic polymer beads is the high pH-stability compared to other MELDI materials such as cellulose or silica particles. As a consequence, elution can also be performed at very high or low pH values.
Furthermore, the MELDI carriers do not affect the spectral patterns in terms of background signals (except most nanomaterials below 1000 Da). Perfect automation can be reached for spherical cellulose beads, poly(GMA/DVB) beads and modified silica particles, whereas liquid handling of nanomaterials by robotics still is difficult to realize. The ongoing mass profiling of hundreds of samples by MELDI is applied to distinguish between diseased and normal samples at the pattern level. For further identification, bound serum constituents can be extracted from the MELDI material or enzymatically digested on-carrier, followed by LC–MS–MS analysis. Nevertheless, well-designed studies are critical for proper interpretation of results. In particular, sampling has a major influence on the recorded MELDI-profiles, and can thus markedly influence and confound the results of the MS analysis, whereas clinical variables might remain unchanged. Inadequate or incorrect processing can result in data sets that are biased, making it impossible to obtain reliable biological information. For that reason a well-established workflow starting from sampling leading to MS analysis and data interpretation is vital to obtain reliable results.
Douglas Gjerde is the founder of PhyNexus Inc., a private company located in San Jose, California, USA, with automated and miniaturized sample preparation tools for the biological research market. Gjerde founded Sarasep Inc. in 1990 for the commercialization of polymeric separation technologies and co-founded Transgenomic Inc. in 1997 for the commercialization of nucleic acid separation technology. Gjerde holds more than 55 US and European patents and has co-authored six books, two book chapters and more than 40 journal articles in separation science.
Matthias Rainer received his PhD in natural science at the Institute of Analytical Chemistry and Radiochemistry at the University of Innsbruck under the supervision of Professor Günther Bonn. The main focus of his work is to expand the scope of unique and high performance analytical techniques, based on nano- and micro-structured materials such as carbon allotropes, metal-oxides and synthetic polymers. The use of these materials as substrates to specifically bind disease-related proteins from biological samples followed by their mass spectrometric analysis is a major strategy for biomarker discovery.
Chistian Huck is currently an associate professor at the Institute of Analytical Chemistry and Radiochemistry, Leopold-Franzens University (LFU), Innsbruck and is also responsible for scientific collaborations for the LFU with East Asia. From 1995 the main research focus was put on the establishment of novel separation procedures in the fields of metabolomics, phytomics and proteomics. In 1998 the variety of analytical methods was enhanced by infrared (IR) spectroscopic techniques based on near-, mid- and imaging IR techniques. He has been the head of a spectroscopy group at the institute since 2006.
Professor Bonn is the Head of the Institute of Analytical Chemistry and Radiochemistry at the University of Innsbruck. In addition to his academic activities, Bonn also serves as the vice-chairmen of the Austrian Advisory Committee for Research and Technology Development. Bonn has published over 180 papers and more than 20 patents, he has co-authored two books, and possesses global expertise in areas related to the fundamental chemistries and integration processes involved in biomolecular separations, particularly with respect to macromolecules such as nucleic acids and proteins. Günther Bonn did his postdoc at Yale University with Professor Csaba Horvath and is the founder of the Horvath Laboratory of Bioseparation Sciences HLBS in Innsbruck.
Acknowledgments
This work was supported by the Austrian Genome Program (Gen-AU, Vienna, Austria).
References
1. M. Palmblad et al., Proteomics – Clin. Appl., 3(1), 6–17 (2009).
2. Y. Wang et al., Curr. Proteomics, 5(2), 104–114 (2008).
3. A. Jemal et al., CA-Cancer J. Clin., 54(1), 8–29 (2004).
4. J.E. Oesterling et al., J. Urology, 145(5), 907–923 (1991).
5. J. Pannek et al., Semin. Urol. Oncol., 16(3), 100–105 (1998).
6. B. Bogdanov and R.D. Smith, Mass Spectrom. Rev., 24, 168–200 (2005).
7. T. Wehr, LCGC N. Am.,24(9), 1004–1010 (2006).
8. Y. Du et al., J. of Proteome Res., 3(4), 801–806 (2004).
9. S.L. Wu et al., Rapid Comm. Mass Sp., 18(19), 2201–2207 (2004).
10. A. Brock et al., Anal. Chem., 75(14), 3419–3428 (2003).
11. R.C. Dwivedi et al., Anal. Chem., 80(18), 7036–7042 (2008).
12. F. Hillenkamp et al., Anal. Chem., 63(24), 1193–1203 (1991).
13. L.G. Sheffield and J.J. Gavinski, J. Anim. Sci., 81(3), 48–57 (2003).
14. N.L. Anderson and N.G. Anderson, Mol. Cell. Proteomics, 1, 845–867 (2002).
15. C.M. Whitehouse et al., Anal. Chem., 57(3), 675–679 (1985).
16. D. Radulovic et al., Mol. Cell. Proteomics, 3(10), 984–997 (2004).
17. R. Bischoff and T.M. Luider, J. Chromatogr. B, 803(1), 27–40 (2004).
18. L.F. Marvin et al., Clin. Chim. Acta, 337, 11–21 (2003).
19. J. Villanueva et al., J. Clin. Invest., 116(1), 271–284 (2006).
20. I. Feuerstein et al., J. Am. Soc. Mass Spectr., 17(9), 1203–1208 (2006).
21. F.E. Ahmed, Curr. Proteomics, 5(4), 224–252 (2008).
22. C.R. Jimenez et al., Proteomics - Clin. Appl., 1(6), 598–604 (2007).
23. A.K. Callesen et al., Rapid Comm. Mass Sp., 22(3), 291–300 (2008).
24. C.A. Jock et al., BioTechniques, 37(1), 30, 32, 34 (2004).
25. J. Forshed et al., J. Proteome Res., 7(6), 2332–2341 (2008).
26. L.A. Liotta et al., Nature, 425(6961), 905 (2003).
27. L.A. Liotta et al., J. Clin. Invest., 116(1), 26–30 (2006).
28. R.L. Gundry et al., Proteomics — Clin. Appl., 1(1), 73–88 (2007).
29. E.M. Smets et al., Clin. Chem. Lab. Med., 42, 435–439 (2004).
30. A.E. Pelzer et al., BJU international, 99(3), 658–62 (2007).
31. S.K. Drake et al., Clin. Chem., 50(12), 2398–2401 (2004).
32. T.W. Hutchens and T. Yip, Rapid Comm. Mass Sp., 7(7), 576–80 (1993).
33. G. Reddy et al., J. Biomed. Biotechnol., (4), 237–241 (2003).
34. E. Fung et al. Method. Mol. Med., 86, 295–312 (2003).
35. M. Merchant and S.R. Weinberger, Electrophoresis, 21(6), 1164–1177 (2000).
36. C.S. Tan et al., Bioinformatics, 22(12), 1515–1523 (2006).
37. M. Abramovitz and B. Leyland-Jones, Proteome Science, 4, 1–15 (2006).
38. W. Qin et al., PLoS One, 4(1), e4183, (2009).
39. S. Ranganathan et al., J. Neurochem., 95(5), 1461–1471 (2005).
40. H. Zhang et al., Gynecol. Oncol., 102(1), 61–66 (2006).
41. G. Malik et al., Clin. Cancer Res., 11(3), 1073–1085 (2005).
42. E.P. Diamandis, J. Natl. Cancer I., 96(5), 353–356 (2004).
43. E.P. Diamandis and D. van der Merwe, Clin. Cancer Res., 11(3), 963–965 (2005).
44. K.A. Baggerly et al., Bioinformatics,20(5), 777–785 (2004).
45. K.R. Coombes et al., Clin. Chem., 49(10), 1615–1623 (2003).
46. J.L. Luque-Garcia et al., J. Chromatogr. A, 1153(1), 259–276 (2007).
47. A. Jemal et al., Cancer Statistics, CA: a cancer journal for clinicians, 54(1), 8–29 (2004).
48. J.E. Oesterling, The J. Urology, 145(5), 907–923, (1991).
49. I. Feuerstein et al., J. Proteome Res., 4(6), 2320–2326 (2005).
50. M. Rainer et al., Rapid Comm. Mass Sp., 20(19), 2954–2960 (2006).
51. M. Rainer et al., J. Proteome Res., 6(1), 382–386 (2007).
52. L. Trojer et al., Rapid Comm. Mass Sp., 19(22), 3398–3404 (2005).
53. H.W. Kroto et al., Nature, 318, 162–163 (1985).
54. R. Bakry et al., Int. J. Nanomed., 2(4), 639–649 (2007).
55. R.M. Vallant et al., J. Proteome Res., 6(1), 44–53 (2007).
56. L. Trojer et al., J. Chromatogr. A, 1117(1), 56–66 (2006).
57. A. Greiderer et al., Amino Acids, Springer (2008).
58. A.V. Melechko et al., J. Appl. Phys., 97(4), 041301 (2005).
59. M.R. Cuervo et al., J. Chromatogr. A, 1188(2), 264–273 (2008).
60. Z. Guo et al., Anal. Bioanal. Chem., 384(3), 584–592 (2005).
61. S. Iijima, Nature, 354, 56–58 (1991).
62. D.S. Bethune et al., Nature, 363(3), 605–607 (1993).
63. M. Najam-ul-Haq et al., Anal. Chim. Acta, 561(1–2), 32–39 (2006).
64. L.K. Bigelow and M.P. D'Evelyn, Surf. Sci., 500, 986–1004 (2002).
65. X.L. Kong et al., Anal. Chem., 77(1), 259–265 (2005).
Fundamentals of Benchtop GC–MS Data Analysis and Terminology
April 5th 2025In this installment, we will review the fundamental terminology and data analysis principles in benchtop GC–MS. We will compare the three modes of analysis—full scan, extracted ion chromatograms, and selected ion monitoring—and see how each is used for quantitative and quantitative analysis.
Characterizing Plant Polysaccharides Using Size-Exclusion Chromatography
April 4th 2025With green chemistry becoming more standardized, Leena Pitkänen of Aalto University analyzed how useful size-exclusion chromatography (SEC) and asymmetric flow field-flow fractionation (AF4) could be in characterizing plant polysaccharides.
This information is supplementary to the article “Accelerating Monoclonal Antibody Quality Control: The Role of LC–MS in Upstream Bioprocessing”, which was published in the May 2025 issue of Current Trends in Mass Spectrometry.











