Unlocking the Power of Data
LCGC North America
The modern analytical laboratory generates enormous amounts of data. These data are typically stored in vendor-specific, proprietary file formats
The modern analytical laboratory generates enormous amounts of data. These data are typically stored in vendor-specific, proprietary file formats. Historically, the data management problem has been addressed through custom software and patches designed only to mitigate issues without addressing the underlying problem. Here we describe the underlying data-related problems facing the analytical chemistry community and present our collaborative approach to address these problems.

Regardless of the type of measurement, a company's size, or industry sector, analytical data and analysis thereof underpins the fundamental processes and decision making for companies and businesses operating within the analytical community. Arguably, this makes data an asset of indispensable value for any of these companies, on comparable standing with its people and products. Because of the sheer volume and complexity of the data, the management and maintenance of this valuable asset is associated with numerous challenges, which in turn create additional challenges in fully leveraging its value. In practice, significant value remains untapped because the attention and investment today in data management tools and practices are not commensurate with the volume, complexity, and value of our analytical data. Furthermore, when compared to other industry sectors, these challenges are exacerbated for any company that works in domains with additional requirements associated with regulatory compliance, like the pharmaceutical and biotechnology industry. When considering the importance of data in daily business operations, why then is it so common that data management continues to be so difficult, time consuming, and costly - all of which affect the efficiency and subsequent profitability of companies? Can a tacit acceptance of added cost and complexity be best practice or good business? A series of articles in 2010 and 2011 described the problems associated with analytical chemistry data and proposed an approach to address them (1,2). As a direct result of these papers, a group of pharmaceutical and biotechnology companies formed Allotrope Foundation in 2012 to address the paradox between the ease of generating enormous amounts of data and the difficulty in extracting value from these data. The members of the foundation have recognized that through collaboration, sharing of resources, and expertise, it is possible to change the status quo. What follows is a description of the approach taken by Allotrope Foundation and progress toward solving these problems by addressing their root cause. Though initially conceived and initiated in the pharmaceutical industry, there is ample evidence that neither the problems nor the approach to a solution are uniquely defined by the nature of the samples being analyzed. Rather, any company in any industry using electronic data management in analytical chemistry would be subject to the same issues, and realize similar benefits from the work of Allotrope Foundation.
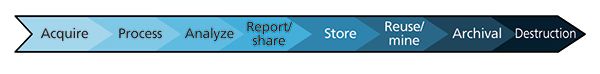
Figure 1: The analytical data life cycle. The data life cycle begins with "data acquisition" and ends with "data archival," followed by its "destruction" in some cases.
The Problem Statement
The pharmaceutical and biotechnology industries generate enormous amounts of data in all aspects of their daily operations such as research and development, and manufacturing. Even considering only the analytical experiments conducted for just one drug product, the volume of data that needs to be managed is enormous. Since the cost of data storage continues to decrease and the speed at which data can be transmitted continues to improve, it is not the quantity of data that is the problem, rather it is the means by which the data are recorded, described, indexed, and stored. The accumulation of small issues or gaps throughout the entire data life cycle (Figure 1) affects the ultimate value or usability of the measured result in a profound way. Like any imperfection in a multistep process, it can be difficult or impossible to find, let alone correct the original mistake, particularly if some information is missing or contradictory. Three fundamental points of failure or complexity are at the root of the problem and are described below: proprietary file formats, inconsistent contextual metadata, and incompatible software. Addressing these fundamental issues will positively affect the ability to use, exchange, transmit, recall, and extract value from data at every stage of their life cycle and address the underlying data management problems facing today's modern analytical laboratory (Figure 2).
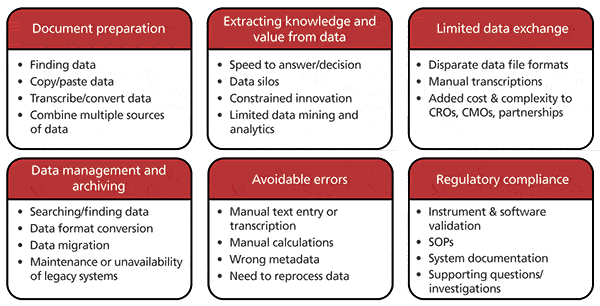
Figure 2: A representative set of downstream effects caused by the current data management problems.
Proprietary File Formats
As the science and associated technologies of analytical chemistry have evolved, the broad landscape of instruments and software applications has generated an equally diverse array of proprietary data formats and systems to consume them. Unfortunately, this diversity in data formats hinders the ability of these systems to consume and share data. The data and associated metadata stored in these proprietary file formats are effectively inaccessible outside of their native vendor application. Proprietary formats pose a serious problem for companies that wish to share data or electronic instrument methods between business units or with external collaborators and partners such as contract research organizations (CROs), particularly if each site or partner utilizes different software or hardware in their analytical workflows. To circumvent this problem, proprietary data and method files are often converted into "compatible" formats for exchange and then are subsequently converted back into the proprietary formats required by the software used by the recipient. For electronic methods, this conversion is routinely accomplished "by hand," in which the contents of a static PDF document are read and manually transcribed by a human into the instrument control software. Furthermore, static PDF documents often do not adequately reference the data or provide the complete metadata package, such as audit trail information, which would exist in the original electronic records. Each of these conversion and manual transcription steps impedes data exchange, analysis, and archival and increases the possibility for the introduction of errors and misinterpretation into the process, especially where manual steps are required.
Besides impeding facile data use and exchange, proprietary file formats also introduce significant long-term retention problems. Like the pharmaceutical industry, many industries need to archive data for regulatory, legal, business, or intellectual property reasons, often for extended periods (such as decades). Continuous and rapid technological evolution in computers, software, and hardware results in inevitable obsolescence, causing incompatibilities to accumulate over time, and in turn results in a significantly diminished ability to use archived data. To allow future access to data stored in and dependent on obsolete and unsupported file formats, companies often maintain museum-like storehouses of decommissioned hardware and software. In addition, companies regularly and on a recurring basis invest enormous resources in data migration projects to periodically move data to current systems, only to have to repeat this exercise a few years later when the "new" software system becomes "old." Nonetheless, all the steps that are undertaken today to provide future readability of archived data only increase the likelihood that these data can be accessed in the future, they do not guarantee it.
Inconsistent Contextual Metadata
Metadata capture the "who," "what," "when," "where," "why," and "how" the data were generated - that is, all the information needed to describe the data and the context in which they exist. While some software systems record certain pieces of metadata automatically, the information captured is typically not sufficient to provide the level of experimental detail required for full interpretation and contextual understanding after-the-fact, nor are these metadata readily available to downstream software applications. As a result, the burden to fill in the metadata gaps falls to the scientist. These metadata gaps are filled through a combination of free-text entry or selections from a limited, predefined set of local vocabularies that are often inconsistent between software applications. By relying on free-text entry, the possibilities of deviating from an agreed vocabulary or format, introducing spelling errors, or just conveying inaccurate information significantly increases the chance of inadvertently recording bad metadata. This combination of free-text errors and limitations of local, predefined vocabularies, coupled to the likelihood of blank metadata fields greatly hinders the usefulness of the metadata that are captured and subsequently the usefulness of the data themselves. Whereas humans can read a variety of spellings and abbreviations of a word or phrase and automatically interpret them to mean the same thing, software-based searching or aggregating data based on this kind of unstructured data is much more difficult, and in most situations requires manual intervention.
Because important metadata, such as instrument methods, settings, and so on, are stored within proprietary formats, the aforementioned problems associated with proprietary formats directly and materially contribute to the reality that contextual metadata are often incomplete, inaccurate, or incorrectly captured throughout the analytical workflow. Given the number and diversity of software applications used in the typical analytical laboratory, pieces of this critical context are spread across multiple software applications and sources, electronic documents, and on paper. The probability of losing the knowledge of where these different pieces of context are stored, and how the local vocabulary and syntax are to be interpreted increases as time passes especially as data are shared and used further from their origin. The loss of this context subsequently erodes the value of the data, until finally it is deemed less trouble to repeat the measurement than find or understand the original data.
Incompatible Software
The analytical laboratory depends on a significant number of software applications including electronic laboratory notebooks (ELN), laboratory information management systems (LIMS), chromatography data systems (CDS), and data analysis packages, just to name a few. In addition, analytical instruments and even the detectors themselves may have stand-alone software required for instrument control, data capture, and data analysis. But, unlike the "app stores" for mobile devices with literally a million applications that can be downloaded and run instantly, the modern analytical laboratory is not a "plug-and-play" environment. It can cost millions of dollars and require a small team of experts to install and validate software (above and beyond the requisite license fees). The patchwork of software that needs to be interconnected to enable the acquisition, transfer, analysis, and storage of data can be numerous and complex. There are several routes available to a company to bridge software applications such as: internal developers using vendor provided software development kits (SDKs) to create adaptors to allow the output from one program to feed into another program; software integration vendors contracted to develop custom software to connect software; or the standardization within a company or department on a single vendor for their analytical laboratory needs. Typically, in practice it is a combination of several solutions used to interconnect software systems, which results in a highly customized and likely expensive undertaking that rarely, if ever, creates a fully integrated data-sharing environment. Furthermore, because of the level of customization, laboratories frequently have to deal with issues arising from software updates and version changes along with the introduction of new technology into the laboratory. The result is a perpetual cycle of creating expensive stop-gap solutions to keep the data flowing.
The Proposed Solution - "The Allotrope Framework"
Companies have historically dealt with the aforementioned data management problem by focusing on and addressing the downstream effects illustrated in Figure 2. The explosion of data coming from analytical laboratories is rendering this position increasingly unsustainable and untenable. These effects may originate in the analytical chemistry laboratory, but are felt throughout the organization, even in those areas that do not have direct or knowing contact with "data." Accordingly, Allotrope Foundation member companies devised a holistic strategy to deal with this problem and to create a solution that addresses the root cause of the problem - the incomplete, inconsistent, and potentially incorrect metadata (captured through manual entry) and the lack of a standard file format to contain the data and associated metadata. This solution forms the basis of the "Allotrope Framework," which is composed of three parts: a nonproprietary data format, metadata definitions and repository, and reusable software components to ease adoption and ensure consistent implementation of the other two components
Standard Data Format for Analytical Chemistry
Given the longevity of record retention and access requirements by regulatory agencies (such as the Food and Drug Administration [FDA], European Medicines Agency [EMA], and Pharmaceuticals and Medical Devices Agency [PMDA]), the Allotrope Framework will include the development of a publicly documented file format for the acquisition and exchange of data that will allow for easier long term storage and access to both the analytical data and associated metadata. As such, the format must not be limited to storing data; it must also be the format to store the context in which the experiment is performed as well as the method used to acquire the data. The ability to create and seamlessly share a vendor-neutral instrument method file eliminates the "PDF file-sharing method" along with the requisite manual transcription steps.
As described above, the readability of analytical data today and far in the future is critically important, with some companies' data retention policies spanning 25, 50, or even 100 years. Allotrope Foundation recognizes that data files created today need to be directly and easily accessible to other software packages, processes, corporate entities, collaborators, and regulators today as well as in the future. This accessibility far into the future will be made possible through the evaluation, selection, federation, and subsequent implementation of today's best-in-breed data standards to create a standardized, obsolescence-resistant, nonproprietary data format. However, it is certain that despite the best intentions of Allotrope Foundation, the selected data standard will need to be updated to newer technology at some point in the future. Making the technical details of the standard format publicly known ensures that third parties could provide solutions to access or migrate the existing body of standard format data, as needed.
To facilitate adoption by the vendor community, the standard data format is being designed from first principles to meet the performance requirements of modern instrumentation and to be readily extensible. Extensibility provides resistance to technological obsolescence by allowing new techniques, combination of techniques, and technologies to be incorporated while maintaining backwards compatibility with previously released versions. This backwards compatibility is especially important when considering the archivability of the file format.
Metadata Definitions and Repository
The ability to easily read data in the future is practically useless without knowing the context around which the data were gathered or even at a more basic level, being able to find the data in the first place. It is the retention of complete, consistent, and correct metadata that allows data to be efficiently searched and retrieved. As such, the capture and storage of all relevant contextual metadata is a necessary precondition to mitigating the data management problem and is critical to an archive-ready file format and data integrity.
The metadata definitions provide the ability to consistently and accurately describe the methods, the experiment, and the context in which they occur. To address the shortcomings in the current metadata capture process, which relies heavily on free-text manual entry, we envision that many manual entry steps can be automated so that standardized, predefined metadata terms are obtained from authenticated sources, initiating their look-up via the barcode of a sample, or near-field communication (NFC) of an employee's badge with password authorization, or triggering a pick-list of values from the software user interface to select appropriate parameters (retrieved from a controlled vocabulary source). This strategy eliminates errors associated with unverified free-text entry since it no longer occurs. It also eliminates inconsistent metadata because scientists are restricted to predefined and approved terms that are relevant to the experiment at hand. In turn, this reduces the burden to record experimental details, including those details unknown or not easily accessible to scientists. The automation is carried one step further in the metadata approach created by the Allotrope Framework, in which metadata stored alongside the data will grow and will be augmented as the data move through the product life cycle in an automated fashion. This allows a "data map" of the laboratory to be created and searched in ways not possible today.
The approved or predefined vocabularies are provided to users through an extensible "metadata repository." The Allotrope Framework will contain integrated metadata repositories containing common or core terms, company-specific terms, and vendor-specific terms. This modular approach to the metadata repository allows for future extension of terms as the underlying science advances, new techniques and technologies become available, or business processes change through evolution, acquisition, or divestiture. In addition, industry-specific metadata repositories can also be created and incorporated into the framework allowing companies outside the pharmaceutical industry to implement the Allotrope Framework.
Reusable Software Components
Although the standard data format and metadata components of the Allotrope Framework are relatively simple and straightforward at a conceptual level, at the level of a practical implementation these components are all highly detailed and complex. As any solution becomes more complex, the barrier to implement and adopt that solution increases dramatically, thus reducing the likelihood of its use. Moreover, consistency is critical - an inconsistently implemented or unused standard of any kind is not a standard. To significantly lessen the complexity associated with implementing the Allotrope solution, while at the same time ensuring consistency of the implementation, a third component is needed: readily available software tools that will allow developers to easily and consistently embed the Allotrope Framework into their software without requiring an underlying, detailed knowledge of the technical aspects of the standards or analytical techniques. By lowering the barrier to implement the standards, the framework provides the means to drive their widespread adoption.
The software tool set provided will consist of application programming interfaces (APIs), class libraries, and associated documentation. This approach is analogous to APIs provided by iOS, Android, and Windows: toolkits that enable applications that allow us to exchange texts, hyperlinks, and images using different mobile devices, all of which use underlying standards that are largely invisible to software developers. Similarly, vendor-provided SDKs can be employed today by end-users to create applications for, or extensions to the vendor software, both of which are external to the original software application. The goal of Allotrope Foundation in providing the Allotrope Framework "toolkit" is to allow vendors to integrate the Allotrope Framework, and the standards it encompasses, directly into their software products. During the early stages of deployment and integration of the Allotrope Framework in everyday use it might be necessary in some cases to use the vendor SDKs to create adaptors to allow the current generation of software (as well as legacy systems) to interface with the Allotrope Framework. We envision that in the future new versions of software will be provided by vendors in an "Allotrope Ready" format, with Allotrope Framework components or APIs embedded directly into their applications and solutions. In this future state, Allotrope Foundation foresees that the Allotrope Framework toolkit will evolve to be a means for vendors, third parties, and end-users to innovate and exploit analytical data and metadata in ways not possible today.
A schematic illustrating a proposed reference architecture integrated with the Allotrope Framework is shown in Figure 3 and at a high level consists of: the laboratory workflow, metadata repository, and data repository or archive. Allotrope Framework APIs and class libraries provide connectivity between and within these high level objects. The laboratory workflow contains all the components including the associated instruments, software, and outsourcing required to execute experiments, acquire data, analyze data, and create reports (additional components that are typically required and are provided through vendor-supplied applications are not illustrated for clarity). In addition, the analytical laboratory requires some form of data storage element. The Allotrope Framework provides the metadata vocabularies and dictionaries to the reference architecture and supplies the means to connect all of the components to one another through the use of the Allotrope Framework APIs and class libraries. The result is a laboratory comprising interconnected components, which can be software or collaborators, all producing and using data in a common format that is able to be readily shared and stored in an archive-ready format. In addition, these data contain complete and accurate contextual metadata provided through the metadata repository, which ensures data integrity and enables data mining capabilities not possible in today's laboratory.
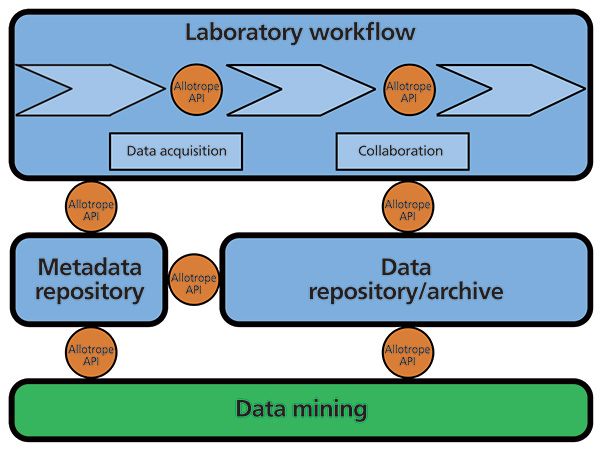
Figure 3: The future analytical laboratory. A schematic illustrating a proposed reference architecture incorporating the Allotrope Framework is shown and consists of: the laboratory workflow, metadata repository, and data repository or archive (blue boxes); the interconnectivity of all the laboratory components and steps within the laboratory workflow (light-blue chevrons) is provided by the Allotrope Framework APIs and class libraries (orange circles). Additional components that are typically required and provided through vendor-supplied applications are not illustrated for clarity. The new level of data mining capability made possible by implementation of the Allotrope Framework and concepts is represented as a green box.
Delivering the Solution
Allotrope Foundation is guided by the principle that the best learning comes from doing. This thinking pervades all aspects of the organization. As part of the Allotrope Framework development, components are first tested through proof-of-concept applications (PoCs) specifically designed to test concepts, approaches, implementation details, and existing standards for data, metadata, and software. This allows for concepts and hypotheses pertaining to the Allotrope Framework to be rapidly tested and refined based on the collective industry expertise from Allotrope Foundation subject matter experts (SMEs), industry best practices, and the vendor community. This approach is consistent with the agile, iterative software development approach being used for the Allotrope Framework and PoC development (Figure 4). The PoCs allow for feedback and input from these various information sources to be readily accommodated into the development process. The extensive use of PoCs and "real-world" testing and evaluation as an integral part of the development process reduces the risk of creating a solution that will have limited practical applicability. In addition, there is a dedicated team tasked to ensure that all Allotrope Framework development throughout the entire software development life cycle (SDLC) is carried out in accordance with our understanding of regulatory and best practice expectations, including appropriate documentation and controls (beginning even with the PoCs described below). Furthermore, the Allotrope Foundation's iterative approach allows for instrument and software vendors, regulatory agencies, and academia to contribute to the development cycle. During the first year of development, the main objective of Allotrope Foundation was to verify the main Framework concepts through the creation and evaluation of three PoCs: instrument software integration, capture of contextual workflow metadata, and archiving support. The Analytical Information Markup Language (AnIML) was used as the initial standard based on its maturity and the fact that it includes concepts from previous standards such as the Joint Committee on Atomic and Molecular Physical data – Data Exchange (JCAMP-DX), Generalized Analytical Markup Language (GAML), mass spectrometric data extensible markup language (mzML), and others (3). These PoCs demonstrate not only the functionality of the framework, but also represent the beginning and end in the data life cycle - data creation and data archiving (Figures 5–7).
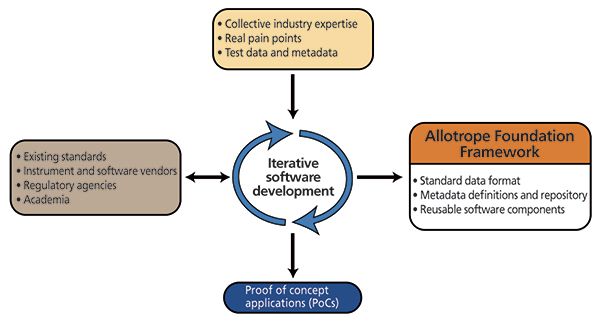
Figure 4: The sources of information utilized in an iterative development approach to PoCs and ultimately the Allotrope Framework components.
Instrument Software Integration PoC
The purpose of the instrument software integration PoC application was to test the ability of a standardized, nonproprietary file format to provide method instructions to native instrument control software from multiple vendors; to use the prototype framework to direct that control software to utilize the supplied method to acquire analytical data on a real analytical instrument; and to store the resulting data back in the standardized format (Figure 5). This PoC demonstrated the ability to create and share methods and provide the resulting data in a standardized format to be read and used regardless of the specific software version or specific vendor used. This concept is rooted in a common real-world scenario - the need to share methods and data between entities, for example, between a sponsor and a CRO.
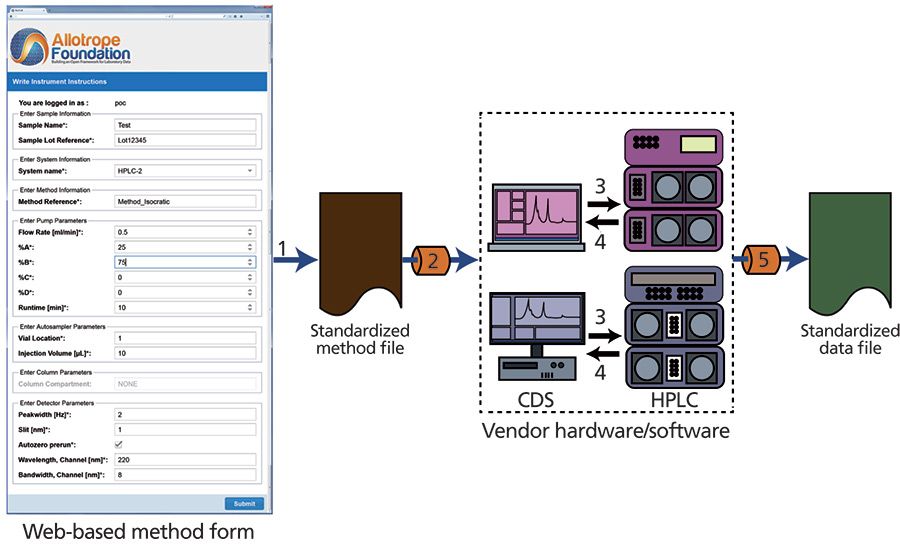
Figure 5: The flow diagram of the instrument software integration PoC. Step 1: HPLC-UV method parameters are input into a web form to create a standardized method file. Step 2: this method file is passed through a vendor-specific adaptor used to convert the standardized instructions into vendor-specific instructions, which are then passed to the CDS for execution. Steps 3 and 4: the CDS executes the run and captures the data. Step 5: the raw data are passed through a second adaptor to convert it into the defined standard format.
For this PoC, high performance liquid chromatography with UV detection (HPLC–UV) was chosen as the analytical technique to be tested because of its ubiquitous use in the laboratory. As mentioned above, AnIML formed the basis of the standardized data format. To demonstrate the vendor-neutrality of the standardized method and data format, the CDS software from both Waters (Empower 3) and Agilent (OpenLAB ChemStation) were used in this PoC. To interact with each vendor's CDS, Allotrope Foundation created adaptors using the vendor SDKs to allow standardized instructions to be sent to each of these CDS platforms and back-convert the CDS-captured data into the standardized data format. The instrument software integration PoC was successfully deployed and implemented at two Allotrope Foundation member companies in early 2014 and subsequently distributed to all Allotrope Foundation member companies for deployment and testing in their individual environments. Future extensions of this PoC will enable the much more complex HPLC–UV experiments that are typically run in modern laboratories, eliminate manual free text entry, and provide a mechanism to support vendor-specific hardware in addition to processed results (for example, peak tables).
Capture of Contextual Workflow Metadata PoC
The ultimate value of data is only realized when the experimental data are associated with accurate and complete contextual metadata. This not only provides the means by which data can be searched and retrieved, it also forms the basis for fueling laboratory automation, data mining, and downstream business intelligence applications. In the metadata capture PoC, a very simple repository containing a standard set of instrument names was created and linked to the instrument software integration PoC. This linkage of a defined dictionary to the web form that was used to define the experiment method allowed the scientist to see the list of available instruments for that experiment, only allowed those instruments to be selected, and ultimately associated the acquired data to that instrument. The metadata capture PoC thus demonstrated the concept to automatically capture metadata provided from a metadata repository and associate these metadata with the original experimental data.
Providing a specific instrument name as standardized, contextual metadata is an illustrative example of the approach the Allotrope Framework is meant to enable, but it clearly doesn't constitute sufficient content to be of any practical utility. As a mechanism to explore which contextual metadata may be most relevant and prioritize its inclusion in the first implementations of the Allotrope Framework, a large body of simulated data was created that included an idealized representation of complete, consistent, and correct contextual metadata for 14,000 analytical experiments representing the typical variety generated in a pharmaceutical company over a decade. This body of simulated data is used to illustrate a variety of ways in which the Allotrope Framework might facilitate the kind of analytics that could deliver insight into a company's data in a way that is a significant challenge and very costly in the current situation.
An illustration of the diversity of such a dataset is presented in Figure 6a. Through the use of suitable analysis tools, the scientist can leverage the complete, consistent, and correct metadata to filter the view, search for specific data, or generate any one of a large number of different aggregate views in seconds as illustrated in Figures 6b–6d. Furthermore, the full integration of the metadata with the experimental data and results will enable facile access to underlying data from the macro-level views. For example, Figure 6e illustrates all the HPLC–UV drug substance stability tests for a given compound, including the ability to visualize individual chromatograms. An impurity threshold can be set interactively to automatically flag individual experiments and by selecting a particular experiment, provide the ability to display the actual HPLC–UV trace. Today, such an analysis using an equivalent amount of real data of a similar broad scope would be exceedingly difficult, laborious, costly, and time-consuming. All of this interactive drill down and data mining in the simulated dataset are achieved with only a few mouse clicks, and hints at just a little of what will be possible once the Allotrope Framework is in widespread use. This detailed level of data mining and analysis is only possible when complete and accurate metadata are preserved with the data and the metadata terms themselves are standardized and predefined across an enterprise.
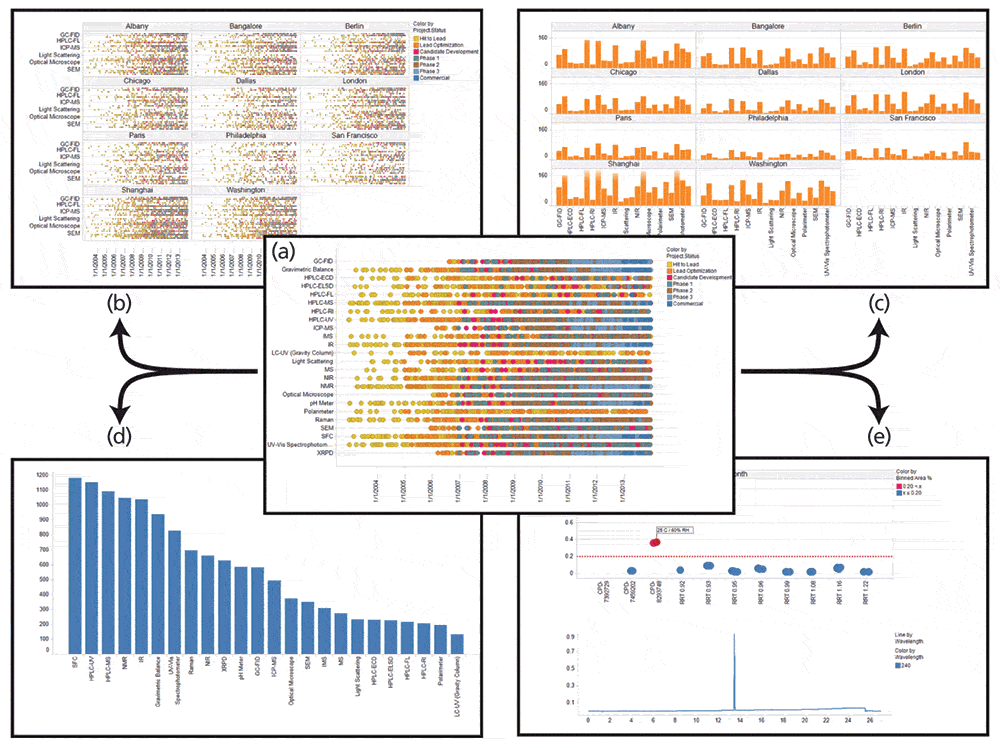
Figure 6: Illustrative data-mining example when complete and accurate metadata are captured. (a) Representation of a simulated dataset that illustrates the diversity of analytical data generated in a typical pharmaceutical company over a 10 year period; Approximately 14,000 simulated analytical experiments are represented, one per dot, organized by technique and color coded according to phase of drug development. (b) All data by site; (c) instrument utilization at each site; (d) instrument utilization globally; and (e) the demonstration of an interactive "drill-down" into the dataset where the selection of any one point in one of the other views displays the details of the corresponding experiment including the data traces.
Archive PoC
As previously mentioned, data are among the most valuable assets of any company. Because the aforementioned problems with contextual metadata are widespread in the current state for many companies, including Allotrope Foundation member companies, the difficulty to find specific data has a profound negative impact on the ability of those companies to preserve and retrieve retained analytical data effectively. This is an acute pain point for Allotrope Foundation member companies, and as a result, the benefits provided by the framework as they relate to long-term data integrity and preservation were deemed an important priority to demonstrate among the first PoC applications.
As such, the purpose of the archive PoC was to demonstrate the ability to store data automatically enriched with the contextual metadata in an archive and to automatically set a retention period based on application of business rules to the contextual metadata associated with the data trace. This PoC also demonstrates the integration with the instrument software integration PoC and metadata repository PoC, as shown in Figure 7, and represents the functional demonstration of three fundamental pieces of the reference architecture described above. As the standardized document containing the acquired data is read into the PoC archive, the contextual metadata contained within it is used to determine the retention period (which itself is stored as an additional piece of contextual metadata in the document), and to index that file so the rich, standardized metadata can now be leveraged for powerful and intuitive searching downstream.
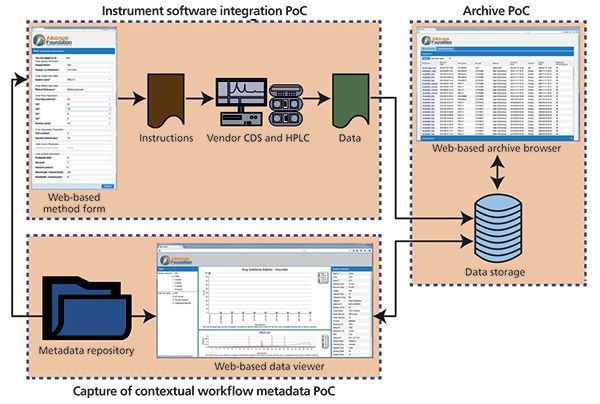
Figure 7: The functional integration of three PoCs demonstrating the potential to impact the data life cycle from data generation through archiving or storage.
Though the PoCs described here are considered "throwaway" applications, all the capabilities described were executed using the underlying framework, which is being developed to a level of quality suitable for commercial use. Furthermore, while the integration of these PoCs represents a vastly simplified version of the analytical laboratory dataflow, it demonstrates the potential utility of the Allotrope Framework throughout the data life cycle (Figure 1) - data generation through archiving and storage. Based on this result, we are confident that all of the intermediate steps between these two endpoints can be enabled via the standard data format, metadata repository, and standard APIs.
Analytical Community Engagement
Allotrope Foundation was created specifically to build the Allotrope Framework and as such, is organized to support the collaborative project team (Figure 8). The current Foundation membership comprises a significant cross section of leading pharmaceutical and biopharmaceutical companies: AbbVie, Amgen, Baxter, Bayer, Biogen Idec, Boehringer Ingelheim, Bristol-Myers Squibb, Eisai, Eli Lilly, Genentech/Roche, GlaxoSmithKline, Merck, and Pfizer. The member companies provide the project funding and SMEs, along with governance and oversight through a Board of Directors while the Consortia Management Team of Drinker Biddle & Reath LLP coordinate and provide legal, scientific, and logistical support. Allotrope Foundation working groups comprised of SMEs focus on defining business requirements and tackling specific tasks or issues, both technical and nontechnical. In addition, Allotrope Foundation has partnered with OSTHUS, a leader in data and systems integration in research and development (R&D) to be the framework architect, working under the direction of Allotrope Foundation with member company SMEs that provide the necessary analytical chemistry knowledge and user requirements. Yet, it is clear that the success of the framework depends on broad adoption and therefore requires the engagement of and input from the greater community of stakeholders. The only way the Allotrope Framework will solve the data management problem of the community is if all the stakeholders in that community contribute to its development.
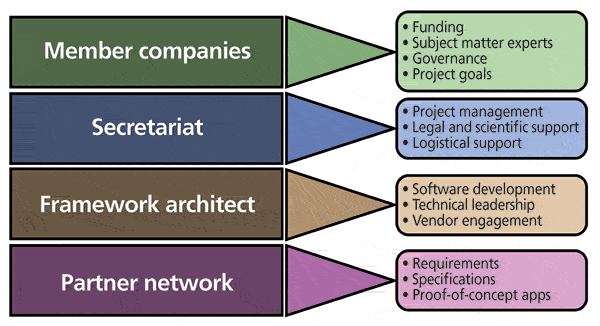
Figure 8: Allotrope Foundation organization. For each of the functional components illustrated, representative roles and responsibilities are shown.
Vendors and Academicians Collaborate Via the Allotrope Partner Network
Although the various industries generating data comprise one dimension of the analytical community, the vendors of the instrumentation and software play a critical role since the easiest path to widespread adoption of the Allotrope Framework (and associated standards) is through directly embedding the Allotrope Framework components into the products they offer. This requires cooperation and collaboration between Allotrope Foundation and the vendor community to ensure that the framework being developed is the right framework and robust enough to accommodate specific vendor needs such as compatibility and performance. Academia represents another community of consumers of instruments and software, but they are also an important source of new methods and technologies driving innovation. To provide a venue for these groups to collaborate in the Allotrope Framework development, the Allotrope Partner Network (APN) (4) was created. The APN program was modeled after similar collaborative programs used by companies to garner input into upcoming products and technologies during the development stage. Members of the partner network benefit from access to common industry requirements, prerelease materials and software, technical support, and can engage directly in collaborative technical work with the Allotrope Foundation project team.
Alignment with Regulatory Agencies
Numerous industries are required to adhere to rules and regulations set forth by governments and enforced by various government regulatory agencies such as the Environmental Protection Agency (EPA) and the FDA in the United States. The ability to find specific analytical data, create reports using those data, and even provide those data upon request to regulatory agencies is a daunting task in the modern analytical laboratory. This is difficult enough with recent data, but is made even more challenging when data from 10 to 20 years ago, or even older, are requested and needed in a short time frame. This challenge is further complicated by the problems associated with reading old data that were acquired on now obsolete technology. Implementing the Allotrope Framework will address these challenges for all future data since the data format will be backwards compatible and always readable. However, to ensure that regulatory agencies agree with the approach taken in creating this framework, Allotrope Foundation welcomes the opportunity to directly engage these agencies and solicit their feedback and support.
Since Allotrope Foundation is primarily made up of pharmaceutical and biotechnology companies, initial outreach efforts have focused on the FDA. In 2010, the FDA Center for Drug Evaluation and Research (CDER) established the CDER Data Standards program and published the CDER Data Standards Strategy Action Plan (5), in which a project entitled "CMC Data Standardization" is described. In addition, the draft of the FDA Strategic Priorities 2014–2018 (6) mentions the FDA's commitment to foster improvements in data management and the use of standards. The goals of Allotrope Foundation are well aligned and consistent with those articulated by the FDA with regard to the data standards strategy and the associated benefits from their adoption and use.
Other Industries Invited to Join Allotrope
Although it was companies from the pharmaceutical and biotechnology industries that formed Allotrope Foundation, it's clear that CROs and contract manufacturing organizations (CMOs) along with other industries such as consumer goods, agrochemical, petrochemical, and environmental health use the same analytical chemistry instrumentation and software within their companies and likely experience challenges similar to those already highlighted above. This was reinforced through recent cross-industry workshops in which representatives from these other industries met with Allotrope Foundation to discuss the Allotrope Framework concept and their analytical data management problems. As a result, membership in Allotrope Foundation is now available to any company that engages in analytical chemistry testing for research, development, or manufacturing of commercial products.
Closing Remarks
As described above, Allotrope Foundation has made tangible progress in creating a sustainable solution to address the analytical data management problems that persist in companies spanning numerous industries. Software development started in September 2013, and in less than one year the project moved from the vision articulated in 2011 to demonstrating that the core concepts of the Allotrope Framework are technically feasible, built functional versions of novel components that did not exist previously, and developed an approach to break a complex problem into tractable pieces. In the process, a deeper understanding has been developed of what it will take to put a version of the Allotrope Framework into production in our companies along with the capabilities exemplified by the PoCs; this will be the subject of the next phase of the project.
Ultimately, what we have learned from the PoCs and our understanding of the requirements to solve this problem is that no one single standard format will meet all the requirements; these technique-specific standards work well for their intended application (for example, mzML for mass spectrometry), but are generally limited in their extensibility for other uses. Furthermore, none of the existing analytical data standards provide an adequate mechanism to preserve the contextual metadata that is as fundamental as the file format itself to solving the problems described above. As a result, the solution provided by the Allotrope Framework will use a federation of existing standards, choosing those best suited for the intended purpose. In addition, the extensible design of the framework allows it to readily accommodate future needs and analytical techniques.
Year Two of Development
The next phase of the project will focus on maturing the established framework and standards, as well as developing the path forward to deliver the framework into production in member companies. Foundation member companies have compiled a list of analytical techniques, software, and initial capabilities required to implement the Allotrope Framework in their laboratories. In addition to furthering the work on HPLC–UV, development for the near term will focus on expanding the list of supported techniques, including mass spectrometry, weighing, and pH measurement. This set of techniques will be supported in the first release of the Allotrope Framework. Numerous standards such as Hierarchical Data Format 5 (HDF5) (7), AniML (8), Batch Markup Language (BatchML) (9), Sensor Model Language (SensorML) (10), and mzML (11) have been systematically evaluated to determine their suitability and ability to accommodate the data and metadata associated with these analytical techniques. As described above, the format will be extensible so as to accommodate additional analytical techniques and standards in future versions.
New PoCs are being defined by Allotrope Foundation and APN members alike that will be developed with collaborations between APN members and the Allotrope and OSTHUS team. A governance model is in place to ensure the PoC process encompasses a wide range of requirements and perspectives, and guide the integration of any subsequent new software components in the Allotrope Framework. Anticipating the availability of the Allotrope and compatible software, Allotrope Foundation members have already begun devising the Allotrope Framework integration and deployment strategy for their individual companies, with projects targeted to begin in 2015.
Invitation to Join Allotrope
Through the participation of a wide and diverse array of analytical community representatives, the Allotrope Framework being developed will be able to meet the individual needs of members of this analytical community. By participating in this endeavor, through membership in Allotrope Foundation or as a member of the APN, you can contribute your expertise toward the development of the Allotrope Framework and help ensure its existence, utility, and quality. The only way to solve an industry-wide problem is to do it industry-wide. Allotrope Foundation invites you to join us and help create this sustainable solution to persistent data management problems.
Acknowledgments
The authors would like to acknowledge the valuable contributions of Allotrope Foundation member company representatives and SMEs, the team at OSTHUS, and members of the Allotrope Foundation Secretariat, all of whom work extremely hard to help ensure the success of this ambitious project while making this a fun and exciting endeavor. We also appreciate advice from the reviewer who provided helpful suggestions to improve the manuscript.
This article was prepared by Allotrope Foundation, which is an association of companies developing a common, open framework for analytical laboratory data and information management. For details, please contact the Secretariat: more.info@allotrope.org.
References
(1) J.M. Roberts, M.F. Bean, W.K. Young, S.R. Cole, and H.E. Weston, Am. Pharm. Rev. 13, 60–67 (2010).
(2) J.M. Roberts, M.F. Bean, C. Bizon, J.C. Hollerton, and W.K. Young, Am. Pharm. Rev.14, 12–21 (2011).
(3) B.A. Schaefer, D. Poetz, and G.W. Kramer, JALA9, 375–381 (2004).
(4) http://partners.allotrope.org.
(6) http://www.fda.gov/downloads/AboutFDA/ReportsManualsForms/Reports/UCM403191.pdf.
(7) http://www.hdfgroup.org/HDF5/.
(8) http://animl.sourceforge.net/.
(9) http://www.mesa.org/en/BatchML.asp.
(10) http://www.opengeospatial.org/standards/sensorml.
(11) http://www.psidev.info/mzml_1_0_0.
James M. Vergis is with Drinker Biddle & Reath LLP and is the Allotrope Foundation Secretariat. Dana E. Vanderwall is with Bristol-Myers Squibb, an Allotrope Foundation Member Company. James M. Roberts is with GlaxoSmithKline, an Allotrope Foundation Member Company. Paul-James Jones is with Boehringer Ingelheim, an Allotrope Foundation Member Company. Direct correspondence to: james.vergis@dbr.com

Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).












