Resolving Volume-Restricted Metabolomics Using Sheathless Capillary Electrophoresis–Mass Spectrometry
Special Issues
Recent advances have significantly improved the performance of capillary electrophoresis–mass spectrometry (CE–MS) for the profiling of polar and charged metabolites in volume-restricted or mass-limited biological samples. Here, those advances are discussed, and attention is also devoted to various technical aspects that still need to be addressed.
The analytical toolbox used in present-day metabolomics encounters difficulties for the analysis of limited amounts of biological samples. Therefore, a significant number of crucial biomedical and clinical questions cannot be addressed by the current metabolomics approach. Capillary electrophoresis–mass spectrometry (CE–MS) has shown considerable potential for the profiling of polar and charged metabolites in volume-restricted or mass-limited biological samples. This article considers advances that significantly improved the performance of CE–MS for in-depth metabolic profiling of limited sample amounts. Attention is also devoted to various technical aspects that still need to be addressed to make CE–MS a viable approach for volume-restricted metabolomics.
A major challenge in bioanalysis and metabolomics is the profiling of (endogenous) metabolites in volume-restricted and often scarce biological samples. Though the analytical techniques currently used in metabolomics are powerful, the need for relatively large amounts of sample for pre-analytics and injection prevent their use in many biomedical and clinical applications. For example, metabolic profiling of small-volume biological samples, such as cerebrospinal fluid (CSF) from mouse models, spheroids or microtissues, liquid biopsies, and samples from microfluidic organ-on-a-chip systems, is seriously hindered by the current analytical technologies because of the limited sample material. The standard analytical techniques also do not allow a maximum amount of biochemical information from valuable and scarcely available biological samples to be obtained because the material is completely consumed for a single metabolomics measurement. Despite important developments in analytical separations technology over recent years, the analysis of small-volume biological samples remains a challenging task. Therefore, there is a critical need for the introduction of analytical methods to allow highly sensitive metabolic profiling of volume-restricted or mass-limited biological samples.
Capillary electrophoresis–mass spectrometry (CE–MS) can be considered an attractive microscale analytical method for metabolomics because in CE nanoliter injection volumes are used from microliter sample amounts or less in the injection vial (1–3). Therefore, CE–MS is highly suited for the profiling of metabolites in ultrasmall biological samples, as has been recently demonstrated for the analysis of CSF from mice and extracts from small tissues or a single cell (4–7). Moreover, CE (denoting here capillary zone electrophoresis [CZE]) separates compounds based on differences in their intrinsic electrophoretic mobility, which is dependent on the charge and size of the analyte and, therefore, well-suited for the analysis of polar and charged metabolites. As the separation mechanism of CE is fundamentally different from chromatographic-based separation techniques, a complementary view on the composition of (endogenous) metabolites present in a given biological sample is provided. In comparison with chromatographic-based methods the separation efficiency of CE is very high because there is no mass transfer between phases, and under perfect experimental conditions the only source of band broadening in CE is from longitudinal diffusion.
Over the past few years, various new CE–MS approaches have been developed that show a strong potential for improving the sensitivity and metabolic coverage in metabolomics (8). This article focuses on advances that significantly improved the analytical performance-especially with regards to improving the metabolic coverage-of CE–MS for volume-restricted metabolomics studies.
Sheathless CE–MS Using a Porous Tip Sprayer
CE is essentially a low-flow microscale separation technique that reaches its optimal separation performance at very low flow rates-typically in the range of 20 nL/min to 100 nL/min-depending on the pH of the separation buffer when using a bare fused-silica capillary. A high separation resolution is obtained in CE by merely separating the compounds based on their electrophoretic mobilities-that is, under (near-) zero electroosmotic flow conditions. The inherently low flow rates of CE are also useful with regards to the electrospray ionization (ESI) mechanism. In ESI, smaller droplets are generated under low flow separation conditions, which results in a more efficient desolvation and an improved transfer of ions to the MS system (9–11). Moreover, at very low flow rates (≤20 nL/min) ion suppression is significantly reduced, resulting in an improved concentration sensitivity (9), which is important for ultratrace detection of metabolites in limited sample amounts.
In a conventional CE system, both ends of the separation capillary are immersed in buffer vials to which electrodes are added to provide a high voltage gradient. To couple CE to MS, the outlet vial must be replaced by an interface to close the electrical circuit and to provide contact with the ESI stream. Therefore, a CE–MS interface needs to apply voltage to the capillary outlet while maintaining independent CE and ESI electrical circuits. A sheath-liquid interface and various other interfacing techniques have been developed to allow the coupling of CE to MS. Until now, most CE–MS-based metabolomics studies have been performed with a sheath-liquid interface (12). CE–MS approaches using a sheath-liquid interface for metabolomics were first developed by Soga and coworkers (13,14). The sheath-liquid interface, originally designed by Smith and coworkers (15), has been used for a broad range of bioanalytical applications with acceptable analytical figures of merit. However, the sheath liquid is generally provided at a flow rate between 2 µL/min to 10 µL/min, thereby significantly diluting the CE effluent and resulting in compromised detection sensitivities for metabolomics applications. Moreover, this flow rate is not compatible with nano-ESI-MS. However, an important benefit of the sheath-liquid interface is that the composition can be modified to improve the ionization efficiency without affecting the selectivity and efficiency of the electrophoretic separation (16,17). The influence of these agents on metabolomics studies by CE–MS still needs to be explored. Overall, as both CE and ESI-MS perform optimally at low flow-rate conditions, the coupling of CE to MS should preferably be performed via an interface that effectively makes use of the intrinsically low flow separation property of CE and the improved ESI efficiency under these conditions.
At present, the development of new or improved interfacing designs for CE–MS and the evaluation of their potential for bioanalysis and metabolomics remains an active area of research (18–25). So far, the most encouraging results for volume-restricted metabolomics studies have been obtained by CE–MS using a sheathless porous tip interface. In this design, which was developed by Moini (19), a porous capillary tip inside a cylindrical metal tube maintains electrical contact via transport of ions and electrons through the capillary wall while spraying the CE effluent into the nano-ESI-MS system. The sheathless porous tip design is especially useful for interfacing narrow (<30 µm internal diameter [i.d.]) capillaries and for low flow-rate (<20–30 nL/min) nano-ESI-MS analyses (26).
In-Depth Metabolic Profiling of Volume-Restricted Samples
The capabilities of CE–MS using a sheathless porous tip interface for global profiling of cationic metabolites in human urine have been assessed (27). Low nanomolar limits of detection (LODs) were obtained for a wide range of metabolite classes in human urine by using only an injection volume of 9 nL. As a follow-up to this work, the potential of sheathless CE–MS for metabolic profiling of very minute amounts of biological samples was assessed. To this end, mouse CSF was selected as a typical example of a biological material only available in minute quantities, that is, only a few microliters can be obtained under proper conditions (4). Figure 1 shows a metabolic profile obtained by sheathless CE–MS for mouse CSF after only a 1:1 dilution with water, thereby fully retaining sample integrity. By using an injection volume of 45 nL from a vial containing only 2 µL of a 1:1 diluted CSF, more than 300 compounds could be observed. As only 45 nL of the sample was consumed, the proposed method allows multiple analyses on a single highly valuable and precious mouse CSF sample, enabling repeatability studies and, even more interesting, analysis of the same sample at different separation conditions to further increase the metabolic coverage. Performing multiple analyses on a single, scarce biological sample is not possible with conventional analytical techniques.
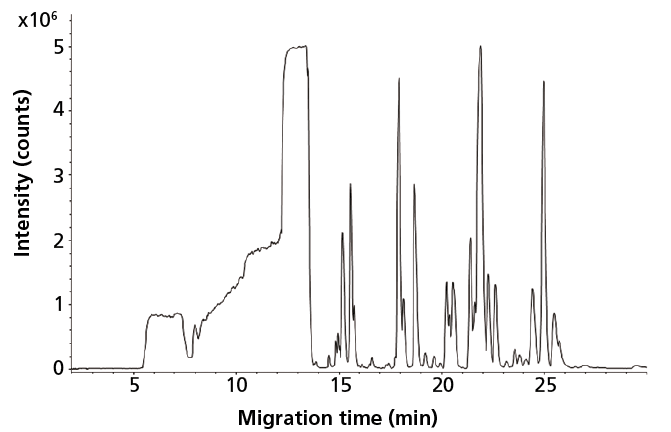
Figure 1: Metabolic profile (m/z 65-1000) of mouse cerebrospinal fluid obtained with sheathless CE-MS using a porous tip sprayer. Conditions: separation buffer: 10% acetic acid (pH 2.2); electrophoretic separation at +30 kV; sample injection: circa 45 nL. Adapted with permission from reference 4.
Apart from volume-restricted biological samples, such as mouse CSF, there is currently a strong interest in analytical tools capable of providing highly sensitive metabolic profiles for microscale cell culture samples. The performance of sheathless CE–MS has therefore been assessed for the profiling of cationic metabolites in an extract from approximately 10,000 HepG2 cells. Figure 2 shows that a reasonable number of metabolites could be detected by sheathless CE–MS using an injection volume of 60 nL, which in this case equals the aliquot of only 5 HepG2 cells. An improvement of the injection part is needed to further enhance the detection sensitivity of the sheathless CE–MS method; however, the results obtained so far clearly indicate the strong potential of the method for metabolic profiling of limited sample amounts.
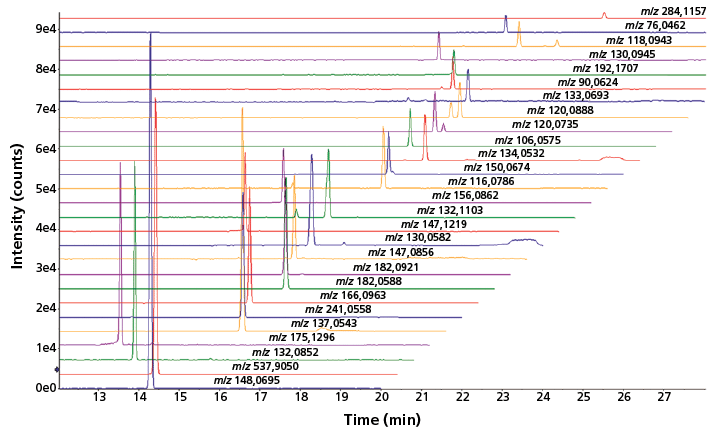
Figure 2: Multiple extraction ion electropherograms for a selected number of metabolites obtained with sheathless CE-MS using a porous tip sprayer in an extract of HepG2 cells. Conditions: separation buffer: 10% acetic acid (pH 2.2); electrophoretic separation at +30 kV; sample injection: circa 60 nL.
Recently, the utility of the sheathless CE–MS method was also examined for the profiling of anionic metabolites in biological samples (28), using the same separation conditions used for the profiling of cationic metabolites-only the MS detection and CE separation voltage polarity were switched. A wide range of anionic metabolite classes could be profiled under these conditions, including sugar phosphates, nucleotides, and organic acids. An injection volume of 20 nL resulted in nanomolar detection limits, which corresponded to a significant enhancement when compared to the micromolar detection limits typically obtained with classical sheath-liquid CE–MS methods. Structural isomers of phosphorylated sugars as well as isobaric metabolites could be selectively analyzed by the proposed sheathless CE–MS method without using any derivatization. The approach was used for the profiling of anionic metabolites in extracts from the glioblastoma cell line. Around 100 compounds were found for an injection volume of 20 nL, corresponding to the content of approximately 400 cells.
Future Directions
The examples highlighted here clearly indicate the ability of sheathless CE–MS using a porous tip sprayer for in-depth metabolic profiling of minute amounts of sample material. Although (sub-)nanomolar detection limits can be obtained for a wide range of polar compounds by only using nanoliter injection volumes, as required for trace-level analysis of metabolites in limited sample amounts, the next important step is to examine the utility of sheathless CE–MS for large-scale volume-restricted metabolomics studies. Hirayama and coworkers have recently shown that a single sheathless porous tip capillary can be used for more than 180 successive runs of a 10-fold diluted human urine sample (29). Still, the long-term performance of sheathless CE–MS for volume-restricted metabolomics needs to be assessed in more extended studies analyzing large numbers of diverse biological samples.
When using a highly sensitive microscale analytical method for in-depth metabolic profiling, attention should also be paid to the design of reliable sampling techniques for volume-restricted or mass-limited samples. Nemes and coworkers have recently developed a microprobe single-cell CE–MS method, which integrates capillary microsampling, microscale metabolite extraction, and CE–MS analysis, for in situ mass spectrometric profiling of metabolites in single cells (7). It is anticipated that microprobe single-cell CE–MS will allow metabolomics studies in larger populations of single cells and other model organisms. Proper sample pretreatment strategies are needed for the selective extraction of polar and charged metabolites from minute amounts of sample. In this context, it would be interesting to evaluate strategies like in-line or on-line solid-phase extraction (SPE)-CE–MS or to explore the possibilities of electrodriven sample pretreatment techniques, such as electromembrane extraction or three-phase electroextraction (30,31). Overall, further developments in this area may result in a more viable CE–MS approach for probing the polar metabolome in volume-restricted samples.
Acknowledgments
Rawi Ramautar would like to acknowledge the financial support of the Veni grant scheme of the Netherlands Organization for Scientific Research (NWO Veni 722.013.008).
Conflict of Interest
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
(1) W. Zhang, T. Hankemeier, and R. Ramautar, Curr. Opin. Biotechnol. 43, 1–7 (2017).
(2) N.L. Kuehnbaum and P. Britz-McKibbin, Chem. Rev. 113, 2437–2468 (2013).
(3) A. Slampova and P. Kuban, J. Chromatogr. A 1497, 164–171 (2017).
(4) R. Ramautar et al., Anal. Bioanal. Chem. 404, 2895–2900 (2012).
(5) J.X. Liu, J.T. Aerts, S.S. Rubakhin, X.X. Zhang, and J.V. Sweedler, Analyst 139, 5835–5842 (2014).
(6) R.M. Onjiko, S.A. Moody, and P. Nemes, Proc. Natl. Acad. Sci. USA 112, 6545–6550 (2015).
(7) R.M. Onjiko, E.P. Portero, S.A. Moody, and P. Nemes, Anal Chem., in press (2017).
(8) P.W. Lindenburg, R. Haselberg, G. Rozing, and R. Ramautar, Chromatographia 78, 367–377 (2015).
(9) A. Schmidt, M. Karas, and T. Dulcks, J. Am. Soc. Mass Spectrom. 14, 492–500 (2003).
(10) M. Wilm and M. Mann, Anal. Chem. 68, 1–8 (1996).
(11) G.A. Valaskovic, N.L. Kelleher, and F.W. McLafferty, Science 273, 1199–1202 (1996).
(12) A. Hirayama, M. Wakayama, and T. Soga, TrAC, Trends Anal. Chem. 61, 215–222 (2014).
(13) T. Soga, Y. Ohashi, Y. Ueno, H. Naraoka, M. Tomita, and T. Nishioka, J. Proteome Res. 2, 488–494 (2003).
(14) T. Soga, Y. Ueno, H. Naraoka, Y. Ohashi, M. Tomita, and T. Nishioka, Anal. Chem. 74, 2233–2239 (2002).
(15) R.D. Smith, C.J. Barinaga, and H.R. Udseth, Anal. Chem. 60, 1948–1952 (1988).
(16) T.J. Causon, L. Maringer, W. Buchberger, and C.W. Klampfl, J. Chromatogr. A 1343, 182–187 (2014).
(17) G. Bonvin, S. Rudaz, and J. Schappler, Anal. Chim. Acta 813, 97–105 (2014).
(18) E.J. Maxwell, X. Zhong, H. Zhang, N. van Zeijl, and D.D. Chen, Electrophoresis 31, 1130–1137 (2010).
(19) M. Moini, Anal. Chem . 79, 4241–4246 (2007).
(20) R. Wojcik, O.O. Dada, M. Sadilek, and N.J. Dovichi, Rapid Commun. Mass Spectrom. 24, 2554–2560 (2010).
(21) X. Guo, T.L. Fillmore, Y. Gao, and K. Tang, Anal. Chem. 88, 4418–4425 (2016).
(22) S.B. Choi, M. Zamarbide, M.C. Manzini, and P. Nemes, J. Am. Soc. Mass Spectrom. 28, 597–607 (2017).
(23) T.T. Nguyen, N.J. Petersen, and K.D. Rand, Anal. Chim. Acta 936, 157–167 (2016).
(24) V. Gonzalez-Ruiz, S. Codesido, J. Far, S. Rudaz, and J. Schappler, Electrophoresis 37, 936–946 (2016).
(25) J. Krenkova, K. Kleparnik, J. Grym, J. Luksch, and F. Foret, Electrophoresis 37, 414–417 (2016).
(26) J.M. Busnel et al., Anal. Chem. 82, 9476–9483 (2010).
(27) R. Ramautar, J.M. Busnel, A.M. Deelder, and O.A. Mayboroda, Anal. Chem. 84, 885–892 (2012).
(28) C. Gulersonmez, S. Lock, T. Hankemeier, and R. Ramautar, Electrophoresis 37, 1007–1014 (2016).
(29) A. Hirayama, M. Tomita, and T. Soga, Analyst 137, 5026–5033 (2012).
(30) A. Gjelstad and K.F. Seip, Bioanalysis 7, 2133–2134 (2015).
(31) R.J. Raterink, P.W. Lindenburg, R.J. Vreeken, and T. Hankemeier, Anal. Chem. 85, 7762–7768 (2013).
Rawi Ramautar studied both pharmacochemistry and analytical sciences at the Vrije Universiteit of Amsterdam, The Netherlands. In 2010, he completed his PhD on the development of capillary electrophoresis–mass spectrometry methods for metabolomics at Utrecht University, The Netherlands. Intrigued by metabolomics for disease prediction and diagnosis, Rawi switched to the Leiden University Medical Center, The Netherlands, to broaden his horizon on this topic. Currently, he is a principal investigator (tenured) at the Leiden Academic Center for Drug Research of the Leiden University where his group is developing microscale analytical workflows for volume-restricted biomedical problems.

New Method Explored for the Detection of CECs in Crops Irrigated with Contaminated Water
April 30th 2025This new study presents a validated QuEChERS–LC-MS/MS method for detecting eight persistent, mobile, and toxic substances in escarole, tomatoes, and tomato leaves irrigated with contaminated water.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)







