“Omics” Applications of Electrochemistry Coupled to Mass Spectrometry — A Review
LCGC Europe
Redox reactions are integral parts of many cellular processes. They are therefore extensively studied in vitro and in vivo. Electrochemistry (EC) represents a purely instrumental approach to characterize direct and indirect effects of redox reactions on bioorganic molecules. This review highlights important trends and recent developments.
Redox reactions are integral parts of many cellular processes. They are therefore extensively studied in vitro and in vivo . Electrochemistry (EC) represents a purely instrumental approach to characterize direct and indirect effects of redox reactions on bioorganic molecules, such as peptides, proteins, and endogenous and exogenous small molecules as well as nucleic acids. In addition to direct infusion electrospray ionization (ESI)–mass spectrometry (MS), hyphenated techniques such as liquid chromatography–mass spectrometry (LC–MS) are often applied for comprehensive characterization of reaction mixtures generated by EC experiments. EC–LC–MS represents a fast, automatable, and "green" approach that can be used in a variety of "omics" disciplines. This review highlights important trends and recent developments.
Redox reactions are essential for life. There are two important groups of biologically important redox reactions: enzymatic and non-enzymatic reactions. Enzymatic redox reactions often involve complex mechanisms of several enzymes. The electrons are transported by flavin- or heme-containing coenzymes from one reaction to another. These reactions represent an integral part of many metabolic pathways. Biological energy, for instance, is stored and released by means of redox reactions. Cellular respiration is the oxidation of glucose to carbon dioxide and the reduction of oxygen to water; photosynthesis involves the reduction of carbon dioxide into sugars and the oxidation of water into molecular oxygen. Furthermore, oxidoreductases such as the members of the cytochrome P450 superfamily generate structural complexity during natural product biosynthesis, and they play a central role in the biotransformation of drugs and toxins ("phase I metabolisms"). Biotransformation reactions involving xenobiotics are usually intended to make them more polar and, thus, enable a more rapid excretion. Biotranformation can lead to loss or gain of activity. Furthermore, in some cases reactive intermediates are produced that can bind to proteins, lipids, and nucleic acids giving rise to cellular damage.

PHOTO CREDIT: HONG LI/GETTY IMAGES
Biologically important redox reactions can also involve nonenzymatic processes. Reactive oxygen species (ROS) are natural by-products of the normal metabolism of oxygen. However, during times of environmental stress (for example, UV or heat exposure), ROS levels can increase dramatically. The availability of ROS can be of importance for an organism, because they are used by the immune system as a way to attack and kill pathogens. Furthermore, some ROS function as physiological regulators of intracellular signalling pathways. In the majority of cases, however, ROS production is considered to be harmful. ROS can react with proteins, lipids, and nucleic acids, which usually gives rise to cell damage called oxidative stress. In humans oxidative stress is thought to be involved in the pathogenesis of different kinds of disease, including neurodegenerative disease, cardiovascular disease, and cancer.
Because of their importance, biological redox reactions are extensively studied in in vitro and in vivo models. Electrochemistry (EC) was found to be a powerful approach to complement existing assays in characterizing direct and indirect effects of redox reactions on bioorganic molecules. EC was particularly useful in generating oxidation products as well as reactive intermediates, which can be trapped by different kinds of electrophiles and nucleophiles. Moreover, EC is a purely instrumental approach. Experimental conditions, including the electrochemical potential, the electrode material, pH, as well as the kind and concentration of reactants, can be precisely controlled. In addition, the use of EC means costly and often non-specific enzymes and the use of harsh chemicals can be made obsolete (1–17). Thus, EC can be regarded as a green or sustainable chemistry approach.
Redox reactions may give rise to the formation of complex mixtures of intermediates, products, and by-products. Different kinds of analytical techniques can be used for comprehensive analysis. Mass spectrometry (MS) in particular has found widespread application for that purpose. Mass spectrometric techniques enable qualitative (that is, identification and structure elucidation) and quantitative analysis. Among the different ionization techniques available, electrospray ionization (ESI) is the most commonly applied technique. ESI allows the MS characterization of a large variety of compounds ranging from small molecules to large biopolymers. The importance of EC–ESI–MS techniques is emphasized by the large number of reviews that have been published in recent years (1–17). Particularly in life sciences, EC–ESI–MS has found widespread application. This review will give a short overview on recent advances in that important field of research (Figure 1).
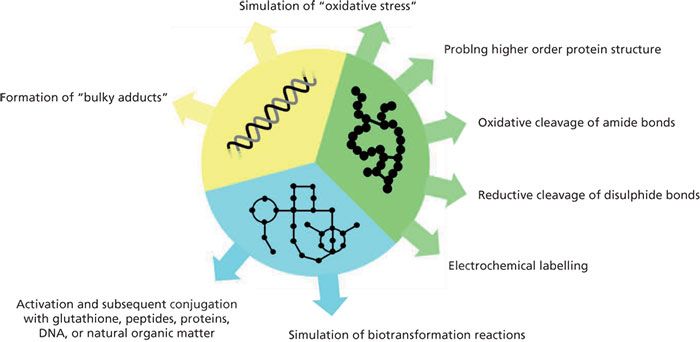
Figure 1: Sketch summarizing the different kinds of applications that are realizable with EC–(LC)–ESI–MS in metabolomics, proteomics, and nucleic acids research.
Experimental Setup
Different experimental setups have been developed that enable EC–ESI–MS experiments. In the simplest setup, the inherent EC of ESI is used (14). From the electrochemical point of view (18–20), the ESI source represents a controlled-current cell consisting of two electrodes. One electrode is the capillary emitter; the mass spectrometer acts as the counter electrode. The two electrodes are connected on the one hand by the power supply and on the other by a series of resistors consisting of the electrochemical contact to the solution, the solution resistance, the resistance at the solution-air interface and in the gas-phase, and the charge neutralization at the counter electrode. During operation, charges are transported from the emitter via the gas phase to the mass spectrometer. The loss of charges needs to be balanced in solution, and this is accomplished by electrochemical processes at the emitter electrode. These redox reactions may involve analytes (21–23), the solvent (24,25), or the electrode material (18,26). Practitioners have learned to control the electrochemical part of ESI in a way that the impact of EC on the mass spectra observed can be very much tuned. In the majority of cases, experimental conditions are chosen that prevent the mass spectrometric detection of ESI-inherent electrochemical reactions. However, in some situations EC can be used as an analytical advantage. A clear limitation of this setup is its inability to precisely control the electrochemical potential at the emitter electrode. Thus, experimental setups allowing separation of the electrochemical processes studied from the ESI-inherent EC are more commonly used.
Discrete electrochemical cells can be on-line hyphenated to ESI–MS (Figure 2[a]). The electrochemical cells used are typically controlled-potential cells consisting of three electrodes (that is, working electrode, reference electrode, and auxiliary electrode). The cells contain either porous flow-through or planar, thin-layer, flow-by working electrodes. Cells with porous electrodes are considered to provide good conversion rates even at high flow rates because of the large surface area provided. Typically, glassy carbon is used as electrode material. A clear disadvantage of glassy carbon is the occurrence of analyte adsorption on the electrode surface. Thus, the cells are usually operated with solvents containing high organic contents. Other problems of this cell type are limited robustness and reproducibility. Life history or age of the electrochemical cell can sometimes have an impact on the oxidation reactions observed (27). For the planar thin-layer cells, adsorption on the working electrode is usually a less common problem (28). Another advantage of this cell type is the possibility to use different kinds of electrode materials, such as glassy carbon, boron-doped diamond, platinum, and titanium. For obtaining high conversion rates, thin-layer cells are usually operated at very low flow rates (<10 μL/min) (29).
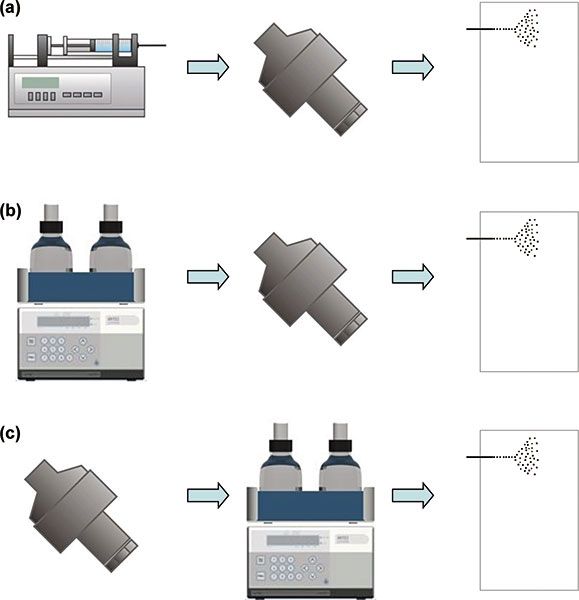
Figure 2: Commonly used setups for combining EC with LC and ESI–MS: (a) EC–MS, (b) LC–EC–MS, and (c) EC–LC–MS.
Electrochemical cells integrated in microfluidic devices were presented (30–35). With this design, high electrochemical conversion efficiency can be obtained with only small amounts of reagent being used. Another advantage of such a setup is the short transit time between EC and MS, which enables the detection of short-lived oxidation products (35).
Configurations with electrochemical cells integrated into the ESI emitter are available (36–39). Very recently, we demonstrated that such devices can characterize the reducing potencies of natural antioxidants such as polyphenols in wine (39). The ESI source represents a controlled-current electrochemical cell. The interfacial potential at the emitter electrode is at or near the electrochemical potential of those reactions that sufficiently supply all the required current for the ESI circuit. Indicator molecules prone to oxidation in ESI, such as amodiaquine, can be used to visualize the impact of reducing compounds on the interfacial potential. The extent of inhibition of the oxidation of the indicator molecule is dependent on the kind and amount of antioxidant added.
The coupling of a discrete electrochemical cell with the ESI–MS system is complicated by the need to decouple the cell voltage from the ESI high voltage (40). Decoupling is necessary for proper control of the electrochemical potential at the working electrode, and is usually accomplished either by using sufficiently long transfer lines (~30 cm) between the electrochemical cell and the ESI source (19,36), the insertion of a ground point between the electrochemical cell and the ESI source (36), or by allowing the whole system to float on the potential induced by the ESI high voltage (36).
For proper setup of an EC–ESI–MS system, sites for electrochemical reactions with limited user control should be kept to a minimum. Unwanted secondary reactions can take place at ground points, the ESI source, and auxiliary and reference electrodes (40–42). The mass transport to these sites is usually limited by using high solution flow rates, minimizing their accessible surface area, or avoiding any direct contact with the analyte in the flow stream (for example, separate electrode compartments).
The analytical power of EC–ESI–MS can be increased by integrating liquid chromatography (LC) as an additional dimension of separation. Chromatographic separation is particularly useful to reduce the complexity of the sample submitted either to the EC cell or to ESI–MS. Because of its compatibility with EC and ESI, reversed-phase chromatography is the preferred chromatographic mode of operation. The electrochemical cell can be integrated at different positions into the LC–MS system.
Post-column EC (Figure 2[b]) can be used to study the redox chemistry of selected compounds within complex mixtures (43,44). Pre-column EC (Figure 2[c]) enables the comprehensive characterization of redox reaction products via LC fractionation and subsequent MS detection (45,46). As the commonly applied chromatographic systems are operated at flow rates of several hundred microlitres per minute, electrochemical cells with porous flow-through working electrodes with very high conversion efficiencies are exclusively used for both types of experimental setups. To enable the use of thin-layer cells, the EC cell can be integrated into the injection device (29,47). In such a setup, the sample solution is delivered through the electrochemical cell into the injection loop and subsequently transferred into the LC flow path.
Despite considerable success of the currently available EC instrumentation in converting bioorganic molecules, there is still a need for improved experimental conditions. A lot of research is focused on increasing predictability, yield, and reproducibility of electrochemical reactions. The yield can be improved by optimization of reaction parameters including solvent composition, pH, and electrode material (29). Another strategy to increase the rate of conversion is based on the application of square-wave potential pulses (48,49). The superior performance with pulsing was attributed to an increased desorption of reaction products and continuous renewal of the electrode surface. The potential pulses were also found to give rise to the formation of products that were not seen in direct EC alone. Other approaches developed to expand the reactivity of EC include enzyme-modified systems (50) and hydroxyl radical-producing systems (51–54).
Metabolomics, Lipidomics, and Related Applications
Based on the number of published articles, one of the most important fields of application of EC–ESI–MS lies in studying the redox chemistry of small bioorganic molecules. In this context, EC–ESI–MS techniques are particularly useful to mimic biotransformation reactions.
The first attempts at using EC–MS to characterize oxidation processes of organic molecules date back to the pioneering work of Hambitzer and Heitbaum as well as Yost, Brajter-Toth, and colleagues in the late 1980s (55–59). However, Bruins and colleagues were the first who systematically studied the redox reactivity of pharmaceutical compounds with EC–ESI–MS (60,61). Their work laid the foundation for the use of EC–ESI–MS techniques to mimic phase I oxidative reactions in drug metabolism studies. Because valuable information concerning the sensitivity of a substrate towards oxidation can be obtained from EC–ESI–MS experiments, the technique is regarded as an efficient tool in the drug development process that complements existing in vitro and in vivo screening techniques. EC was found to be particularly useful in cases where P450 enzyme catalyzed reactions are supposed to proceed via a mechanism initiated by a one-electron oxidation. Typically, direct EC oxidation successfully mimics benzylic hydroxylation, hydroxylation of aromatic rings containing electron-donating groups, N -dealkylation, S-oxidation, dehydrogenation, and, less efficiently, N -oxidation and O -dealkylation. The range of oxidizable moieties was extended by the application of hydroxyl radical-producing systems (51–54). For instance, with electrochemically assisted Fenton chemistry aliphatic hydroxylation, aromatic hydroxylation, N -oxidation, and O -dealkylation were also efficiently mimicked. Furthermore, combining EC with enzyme-modified systems holds the promise to enable simulation of the full range of biotransformation reactions occurring in vivo (50).
Although EC–MS has been used extensively to simulate oxidation reactions, it can also be applied to mimic reductive metabolism reactions, and this has been demonstrated for nitro aromatics (62).
The applicability of EC–ESI–MS in drug metabolism studies has been demonstrated for a number of important pharmaceutical compounds, including amodiaquine (50, 63–65), diclofenac (66,67), lidocaine (48,53,54), acetaminophen (63), haloperidol (68), troglitazone (69–71), clozapine (63), trimethoprim (63), flunitrazepam (72), clonazepam (72), chlorpromazine (72), zotepine (73,74), acebutolol (75,76), alprenolol (75), albendazole (77), tetrazepam (47), verapamil (78), and galantamin (79).
Phase I metabolism of drug compounds often gives rise to the formation of reactive species that are generally short-lived and unstable. These species can react with lipids, proteins, and nucleic acids giving rise to substantial damage. Within living organisms reactive intermediates are usually detoxified by binding to glutathione (phase II reaction). Reactive species are also produced in EC experiments. For the detection of these compounds, and to mimic phase II metabolism, trapping agents are commonly applied. Appropriate nucleophiles include glutathione (65,80), glutathione derivatives (81), cysteine (82), and proteins (83). These compounds can be added to the sample solution after or before EC. The study of the skin-sensitizing potential of chemicals is a very interesting application of the ability of EC to produce electrophiles that subsequently react with proteins (84–86). Allergic reactions are most often caused by haptenation of proteins. A presupposition for this reaction is the ability of chemical allergens (small organic molecules and their metabolites) to form covalent bonds to nucleophilic sites in proteins (for example, cysteine and lysine). These activation and conjugation reactions can be studied in a well defined environment of selected nucleophiles with EC–ESI–MS, and this has been demonstrated for p-phenylendiamine, eugenol, and isoeugenol.
The study of biotransformation reactions involving pharmaceutical compounds is also gaining importance in environmental research. Xenobiotics are emitted from living organisms after biotransformation. Furthermore, biotransformation reactions may occur in the environment after exposure to terrestrial and aquatic microbial systems. Accordingly, comprehensive risk assessment of any chemical requires a detailed understanding of its different manifestations. Here, EC–ESI–MS can be used to produce putative metabolites as well as their conjugates with natural organic matter (87–91).
Other emerging fields of application for EC–MS-assisted drug metabolism studies are doping analysis (92,93) and forensic toxicology (94–96). There, EC–MS can be used to predict potential human metabolites of illicit drugs that can be used as targets to verify consumption.
In comparison to drug metabolism studies, the application of EC–ESI–MS to study transformation reactions of endogenous compounds is less common. So far, there are only a few publications demonstrating the potential of this approach (45,46,97–100). Nevertheless, we believe that in the near future EC–ESI–MS will gain in importance in this field of research. EC reactions could become particularly useful to produce sufficient quantities of likely metabolites using known metabolites as reactants to enable their structural elucidation and subsequent confirmation in authentic samples by making use of spectral data stored in libraries or similar systems (101,102).
An example of the successful application of EC–ESI–MS to the study of transformation reactions of an endogenous compound is shown in Figure 3. Cholesterol was oxidized in an electrochemical cell, and the produced oxysterols were characterized by LC–MS–MS. Nine different species were unequivocally identified by comparing the mass spectrometric information to data obtained from reference compounds. The identified products were mostly oxidized near the double-bound at the B-ring (and to a smaller extent at the tertiary carbon in position 25), which is in agreement with susceptibility of cholesterol to free radical driven oxidation.
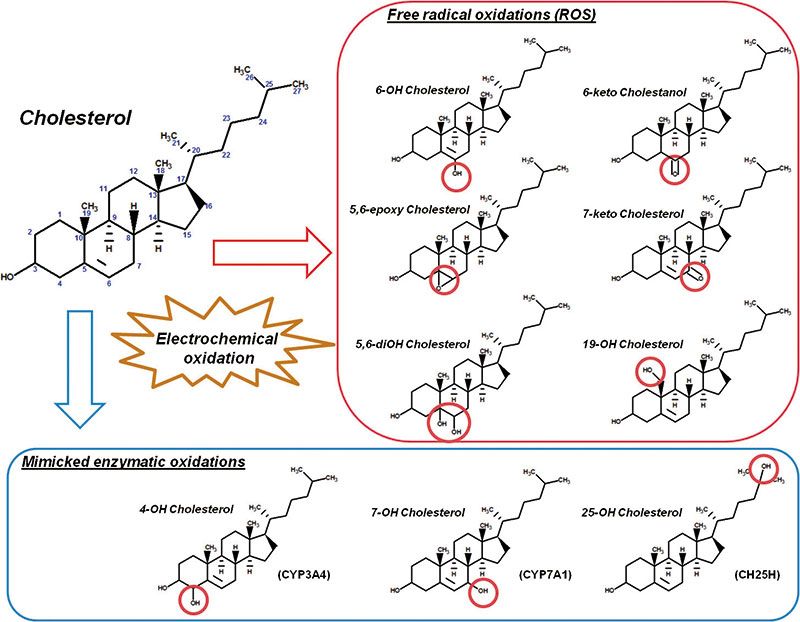
Figure 3: Putative products of electrochemical cholesterol oxidation identified by LC–MS–MS. The carbon numbering at the cholesterol backbone is shown in blue. The sites of oxidation are highlighted by red circles. Indicated are the free radical driven and possible consecutive reactions leading to the formation of the identified compounds, as well as enzymes (in brackets) involved in the generation of oxysterols in vivo. Courtesy of Maria Fedorova and Dieter Weber, Institute of Bioanalytical Chemistry, Faculty of Chemistry and Mineralogy, Universität Leipzig, Leipzig, Germany.
Proteomics Applications
In proteomics, there is a continuously growing interest in EC–ESI–MS. The technique can be used to study the oxidation of peptides and proteins as well as to selectively tag, label, or cleave these species.
Direct oxidation of peptides and proteins is widely studied, because of the role of oxidative damage in disease. The amino acids that most easily undergo oxidation are tyrosine, tryptophan, cysteine, methionine, and histidine (103). Tyrosine and tryptophan are hydroxylated. Cysteine and methionine are converted to their sulphonic acid and sulphone derivatives, respectively. In addition, cysteines can form disulphide bonds, which may be oxidatively cleaved.
The direct oxidation of proteins can be used for analytical advantage to probe their higher order structure by oxidizing solvent-exposed amino acid residues (104,105). Oxidization can be accomplished at physiologically relevant conditions and monitored on-line by MS. A clear drawback of the technique is that, even when only one protein is involved, complications such as low abundances across a range of different oxidized peptides, a wide variety of possible mass shifts, and fragmentation of coeluted oxidized peptide isoforms pose challenges to data interpretation, which currently renders EC more laborious and time-consuming than chemical approaches.
In 2003, Bruins and colleagues realized that direct oxidation of proteins and peptides can induce cleavage of the amide bond at the C-terminal side of tyrosine and tryptophan residues (103). These cleavage reactions were observed in most analyzed peptides and proteins, which clearly suggested that EC could represent an instrumental alternative to chemical and enzymatic cleavage strategies. The main advantage of EC is the direct hyphenation to MS without the need to remove reagents, enzymes, or buffers. Thus, EC–ESI–MS enables direct cleavage and sequencing of proteins. The main problems encountered are related to yield, reproducibility, and adsorption phenomena. The cleavage rate can vary between different species, and a considerable amount of noncleavage oxidation products are often observed (27,106). Nevertheless, the yield of cleavage products can be improved by proper control of the oxidation potential and the application of strongly acidic conditions (pH 2–3) (106). Furthermore, adsorption phenomena and electrode fouling can be reduced by changing from glassy carbon to boron-doped diamond electrodes and by applying regeneration procedures (107).
Direct reduction of peptides mainly targets disulphide bond bridges. Disulphide bonds are one of the most common post-translational modifications and provide covalent cross-linkages in native proteins for maintaining the three-dimensional structures of proteins and their biological activities. However, the presence of disulphide linkages increases the complexity for the protein structure elucidation by MS. Accordingly, they need to be reduced, and this is usually accomplished by chemical reduction. Recently, electrochemical reduction has been shown to represent a fast and efficient alternative for disulphide bond cleavage with immediate application in bottom-up and top-down proteomics (72,108–114). A clear advantage of EC is the possibility to analyze the reaction products directly by MS. EC is a reagent-free cleavage approach. There is no need for any kind of sample preparation between cleavage and MS analysis. High conversion yields can be obtained with proper selection of experimental conditions (that is, Ti-based electrode, 1% formic acid in the solvent). In peptide mapping, EC enables the identification of disulphide-bridged peptides within enzymatic digest mixtures by inducing changes in ion abundance. Furthermore, in combination with tandem MS analysis, disulphide linkage pattern and sequence information for the examined peptides can be determined. At the protein level, electrochemical reduction of disulphide bonds enables tandem MS sequencing with high sequence coverage. For instance, the van der Burgt group reported sequence coverage of over 80% for oxytocin and hepcidin after electrochemical reduction (111). In the case of hepcidin, 21 of the 24 peptide bonds were cleaved after full EC reduction of the four disulphide bonds, while only seven peptide bonds were fragmented in the native hepcidin. Recently, EC has also been applied for the reduction of large proteins such as monoclonal antibodies (mAbs) (113), thereby generating light and heavy chains with high selectivity (Figure 4). Besides the reduction of the four inter-chain disulphide bonds, it was also possible to reduce most of the intra-chain disulphide bonds.
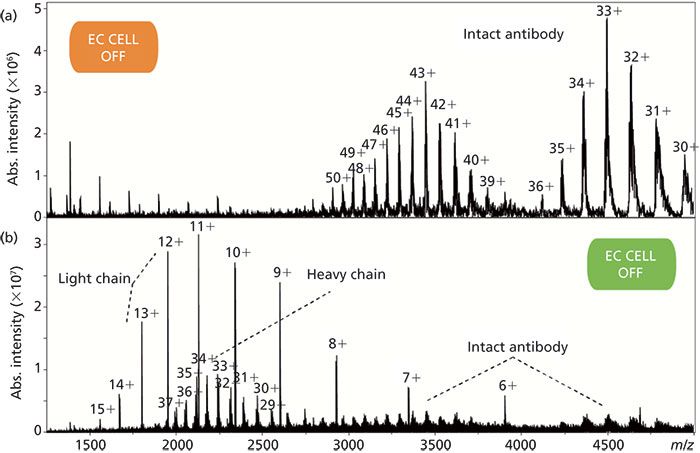
Figure 4: EC reduction of the disulphide bonds of a commercially available monoclonal antibody (mAb) monitored by on-line ESI–FTICR–MS. Cleavage of the four inter-disulphide bonds yields two light and two heavy chains. Courtesy of Simone Nicolardi and Yuri E. M. van der Burgt, Leiden University Medical Center (LUMC), Center for Proteomics and Metabolomics, Leiden, The Netherlands.
Another promising field of EC-based cleavage of disulphide bonds is hydrogen/deuterium exchange monitored mass spectrometry (HDXMS) (114). HDXMS is increasingly being used to characterize the dynamic properties of proteins. When a protein is incubated in D2O, the backbone amide hydrogen/deuterium exchange kinetics directly reflect the conformational dynamics of the polypeptide backbone. HDXMS experiments usually involve digestion of proteins. A critical step in sample preparation is the reduction of disulphide bonds. Cleavage has to be performed under cold and acidic conditions where the amide hydrogen exchange reaction is quenched (pH 2.5, 0 °C). In addition, the reduction must be performed as quickly as possible (within a few minutes) to minimize artifactual deuterium loss or gain in the backbone amides. Here, EC is used for the controlled reduction of the disulphide bonds, replacing the chemical reducing agent Tris(2-carboxyethyl)-phosphine (TCEP), which often causes serious adverse effects on H/D exchange, chromatography, and MS.
EC can also indirectly be used for peptide and protein characterization. Mass tags or labels for specific amino acid residues can be generated electrochemically (9,115–119). Usually, the inherent EC of ESI is applied for activation. Metal ions are used for non-covalent labelling, copper ions for targeting cysteine residues, and zinc ions for targeting histidine residues and phosphorylation sites. Covalent labelling usually involves activated hydroquinone species, which selectively react with cysteine residues. Electrochemically-assisted labelling was found to be useful to determine the number of cysteine residues, which improves protein identification by database searching. Furthermore, based on the labelling extent of different residues within peptide and proteins, the accessibility, reactivity, or affinity of these sites can be tested. The usefulness of this approach has, for instance, been demonstrated by identifying the preferred binding sites of copper ions to β-amyloid 16, which is a protein that is involved in the development of Alzheimer's disease (119).
Genomics Applications
The conical nucleosides - adenosine, cytidine, guanosine, uridine, and thymidine - are the main building blocks of all naturally- occurring nucleic acid polymers (DNA and RNA). Nevertheless, a large number of nucleoside derivatives have been identified in DNA and RNA of living organisms, as well as of viruses, mitochondria, and chloroplasts. The variety of modification reactions is enormous; often oxidation reactions are involved either as part of enzymatic or non-enzymatic processes.
Non-enzymatic oxidation reactions are typically involved in processes leading to DNA damage. Oxidative stress can lead to the production of ROS, which react with nucleic acids. Furthermore, exogenous chemical agents, such as toxins, pharmaceuticals, or pollutants, are often activated by oxidation to react with the genetic material. Produced lesions include base and sugar damages, strand breaks, and crosslinks with proteins as well as the formation of bulky adducts. The cellular response to damage involves several processes, such as DNA repair, cell cycle arrest, and apoptosis, while irreversible mutations contribute to oncogenesis.
To enable the development of strategies to protect the genetic material from oxidative stress, mechanistic aspects of nucleic acids oxidation have been extensively studied. The electroactivity of nucleic acids was discovered by Palecek in the 1960s (120). In the following years, a number of oxidation products of nucleobases, nucleosides, and nucleotides were identified using EC combined off-line with analytical techniques, such as UV–vis spectroscopy, gas chromatography (GC)–MS after derivatization, or LC–MS (121–123). In this context, EC–ESI–MS holds the promise to facilitate and accelerate the identification process by eliminating laborious and time-consuming isolation and derivatization steps (29,99,124–129).
EC–ESI–MS was applied to study oxidation of guanine-containing species. Guanine exhibits the lowest oxidation potential among nucleic acids components and is, therefore, the preferential target of oxidation within nucleic acids. The primary products of guanine oxidation were identified as 8-hydroxyguanine and cross-linked guanines. The product 8-hydroxyguanine represents the most important biomarker to indicate oxidative damage of genetic material, and the formation of this compound clearly demonstrates that EC is able to mimic in vivo oxidation of nucleic acids induced by oxidative stress.
The formation of bulky adducts is an alternative mode of nucleic acids alteration (130). Here, oxidation reactions are involved in activation processes; the produced electrophiles react with nucleophilic sites within the genetic material. EC–MS represented a useful tool to study the formation of adducts between nucleic acids and environmental pollutants (131), amino acids (132), and pharmaceutical compounds (124,125).
The most important enzymatic modification in genomic DNA is methylation of the C5 atom of cytosine (C). Production of 5-methylcytosine (5mC) is catalyzed by DNA methyltransferase enzymes. Methylation of C is an important epigenetic DNA modification that is essential for development and normal function in all mammals (133). In 2009 it was demonstrated that genomic 5mC is transformed to 5-(hydroxymethyl)cytosine (5hmC) (134,135). Oxidation was accomplished by the ten-eleven translocation enzymes. These iron and 2-oxoglutarate-dependent enzymes were furthermore able to convert 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (136). The later oxidation products are considered to represent important intermediates in active demethylation of 5mC. They are recognized and excized by the mammalian thymine-DNA glycosylase, and subsequently converted to C through base excision repair (137,138). There is increasing evidence that 5hmC may also serve as a stable epigenetic mark that possesses unique regulatory functions (139,140). It binds to specific regulatory proteins and is mainly present at actively transcribed genes (141,142).
In vitro experiments employing different sources of radicals demonstrated that 5hmC and 5fC can also be formed by one-electron oxidation of 5mC (143,144). However, both species were considered to represent only minor products of non-enzymatic 5mC oxidation (145). The 5,6-double bond was considered to be more reactive than the methyl group. Thus, oxidation should mainly lead to 5,6-dihydroxy-5,6-dihydro-5-methylcytosine (glycolmC).
However, there is convincing evidence from studying the electrochemical oxidation of cytidine and 5-methylcytidine that 5hmC and 5fC are major products of 5mC oxidation (Figure 5)(126). Simulation experiments have further revealed that C5-methylation reduces stability and increases reactivity of pyrimidine bases. Based on these results, it can be speculated that oxidative stress may induce epigenetic alterations by influencing the previoualy described equilibrium of the oxidized forms of 5mC.
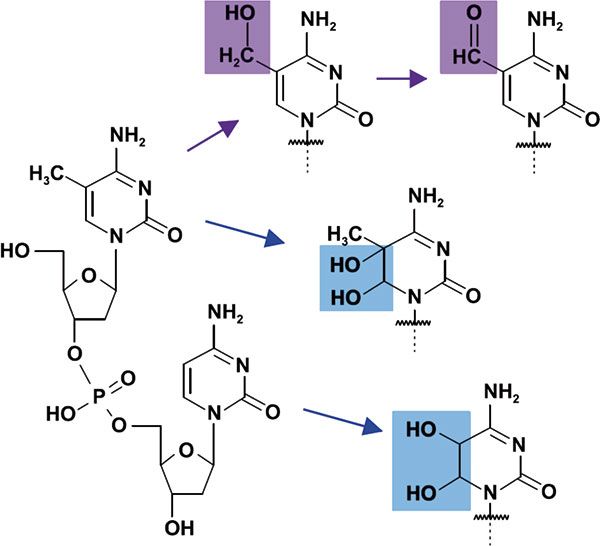
Figure 5: Products of the electrochemical oxidation of C and 5mC.
Conclusions
EC–ESI–MS represents a versatile tool for studying redox processes involving different kinds of bioorganic molecules, such as peptides, proteins, endogenous, and exogenous small molecules as well as nucleic acids. EC–ESI–MS is a purely instrumental approach. EC enables the fast, automated, and cost-effective generation of redox reaction products, and is a "green chemistry" technique. EC–ESI–MS, often in combination with LC separation, allows the comprehensive characterization of the reaction mixture obtained. EC–ESI–MS represents a very useful complement to in vitro and in vivo techniques in metabolomics, proteomics, genomics, and related areas of application. Despite the applicability of EC in general and EC–MS in particular to "omics" this approach is relatively new and therefore unknown to analytical chemists. Nevertheless, the fast growing number of EC–MS publications illustrates the power of this technology and will help to create a broader acceptance and use in life science applications.
Acknowledgements
This work was funded by the Austrian Science Fund (FWF): P 22526-B11. Furthermore, the authors want to thank Maria Fedorova and Dieter Weber (Institute of Bioanalytical Chemistry, Faculty of Chemistry and Mineralogy, Universität Leipzig, Leipzig, Germany) as well as Simone Nicolardi and Yuri E. M. van der Burgt (Leiden University Medical Center (LUMC), Center for Proteomics and Metabolomics, Leiden, The Netherlands) for providing Figures 3 and 4.
Herbert Oberacher is associate professor for bioanalytical chemistry at the Medical University of Innsbruck (Innsbruck, Austria). His research focuses on the development and application of analytical techniques for the analysis of bioorganic molecules with special emphasis on nucleic acids and small molecules. Please direct any correspondence to: herbert.oberacher@i-med.ac.at
Florian Pitterl is research associate at the Institute of Legal Medicine of the Medical University of Innsbruck (Innsbruck, Austria). His main research interests are related to the development and application of LC–MS methods for the qualitative and quantitative analysis of small molecules.
Jean-Pierre Chervet is President and CEO of Antec BV, headquartered in Zoeterwoude, The Netherlands, and with a subsidiary in Boston, USA. His main research interest is the development of electrochemistry–mass spectrometry as a new, sustainable analytical approach.
References
1. H. Oberacher, F. Pitterl, R. Erb, and S. Plattner, Mass. Spectrom. Rev. 34, 64–92 (2015).
2. W. Lohmann and U. Karst, Anal. Bioanal. Chem. 391, 79–96 (2008).
3. U. Karst, Angew. Chem. Int. Ed. Engl. 43, 2476–2478 (2004).
4. G. Diehl and U. Karst, Anal. Bioanal. Chem. 373, 390–398 (2002).
5. S. Jahn and U. Karst, J. Chromatogr. A 1259, 16–49 (2012).
6. H. Faber, S. Jahn, J. Kunnemeyer, H. Simon, D. Melles, M. Vogel, and U. Karst, Angew. Chem. Int. Ed. Engl. 50, A52–A58 (2011).
7. A. Baumann, and U. Karst, Expert Opin. Drug Metab. Toxicol. 6, 715–731 (2010).
8. H. Faber, M. Vogel, and U. Karst, Anal. Chim. Acta 834, 9–21 (2014).
9. C. Roussel, L. Dayon, N. Lion, T.C. Rohner, J. Josserand, J.S. Rossier, H. Jensen, and H.H. Girault, J. Am. Soc. Mass Spectrom. 15, 1767–1779 (2004).
10. H.P. Permentier, A.P. Bruins, and R. Bischoff, Mini-Rev. Med. Chem. 8, 46–56 (2008).
11. E. Nouri-Nigjeh, R. Bischoff, A.P. Bruins, and H.P. Permentier, Curr. Drug Metab. 12, 359–371 (2011).
12. J. Roeser, R. Bischoff, A.P. Bruins, and H.P. Permentier, Anal. Bioanal. Chem. 397, 3441–3455 (2010).
13. J.F. de la Mora, G.J. Van Berkel, C.G. Enke, R.B. Cole, M. Martinez-Sanchez, and J.B. Fenn, J. Mass Spectrom. 35, 939–952 (2000).
14. G.J. Van Berkel and V. Kertesz, Analytical Chemistry 79, 5510–5520 (2007).
15. J. Gun, S. Bharathi, V. Gutkin, D. Rizkov, A. Voloshenko, R. Shelkov, S. Sladkevich, N. Kyi, M. Rona, Y. Wolanov, D. Rizkov, M. Koch, S. Mizrahi, P.V. Pridkhochenko, A. Modestov, and O. Lev, Isr. J. Chem. 50, 360–373 (2010).
16. P.Y. Liu, M. Lu, Q.L. Zheng, Y. Zhang, H.D. Dewald, and H. Chen, Analyst 138, 5519–5539 (2013).
17. U. Bussy and M. Boujtita, Chem. Res. Toxicol. 27, 1652–1668 (2014).
18. A.T. Blades, M.G. Ikonomou, and P. Kebarle, Anal. Chem. 63, 2109–2114 (1991).
19. G.J. Van Berkel and F.M. Zhou, Anal. Chem. 67, 2916–2923 (1995).
20. G.S. Jackson and C.G. Enke, Anal. Chem. 71, 3777–3784 (1999).
21. G.J. Van Berkel, S.A. Mcluckey, and G.L. Glish, Anal. Chem. 63, 1098–1109 (1991).
22. X.M. Xu, S.P. Nolan, and R.B. Cole, Anal. Chem. 66, 119–125 (1994).
23. R. Vessecchi, A.E.M. Crotti, T. Guaratini, P. Colepicolo, S.E. Galembeck, and N.P. Lopes, Mini-Rev. Org. Chem. 4, 75–87 (2007).
24. G.J. Van Berkel, F.M. Zhou, and J.T. Aronson, Int. J. Mass Spectrom. 162, 55–67 (1997).
25. S. Plattner, R. Erb, J.P. Chervet, and H. Oberacher, Anal. Bioanal. Chem. 404, 1571–1579 (2012).
26. G.J. Van Berkel, K.G. Asano, and P.D. Schnier, J. Am. Soc. Mass Spectrom. 12, 853–862 (2001).
27. H.P. Permentier and A.P. Bruins, J. Am. Soc. Mass Spectrom. 15, 1707–1716 (2004).
28. A. Baumann, W. Lohmann, T. Rose, K.C. Ahn, B.D. Hammock, U. Karst, and N.H. Schebb, Drug Metab. Dispos. 38, 2130–2138 (2010).
29. R. Erb, S. Plattner, F. Pitterl, H.J. Brouwer, and H. Oberacher, Electrophoresis 33, 614–621 (2012).
30. V. Mengeaud, O. Bagel, R. Ferrigno, H.H. Girault, and A. Haider, Lab Chip 2, 39–44 (2002).
31. G. Liljegren, A. Dahlin, C. Zettersten, J. Bergquist, and L. Nyholm, Lab Chip 5, 1008–1016 (2005).
32. M. Odijk, A. Baumann, W. Olthuis, A. van den Berg, and U. Karst, Biosens. Bioelectron. 26, 1521–1527 (2010).
33. M. Odijk, A. Baumann, W. Lohmann, F.T.G. van den Brink, W. Olthuis, U. Karst, and A. van den Berg, Lab Chip 9, 1687–1693 (2009).
34. M. Odijk, W. Olthuis, A. van den Berg, L. Qiao, and H. Girault, Anal. Chem. 84, 9176–9183 (2012).
35. M. Odijk, E.J. van der Wouden, W. Olthuis, M.D. Ferrari, E.A. Tolner, A.M.J.M. van den Maagdenberg, and A. van den Berg, Sens. Actuator B Chem. 207, 945–953 (2015).
36. F.M. Zhou and G.J. Van Berkel, Anal. Chem. 67, 3643–3649 (1995).
37. X.M. Xu, W.Z. Lu, and R.B. Cole, Anal. Chem. 68, 4244–4253 (1996).
38. G.J. Van Berkel, K.G. Asano, and M.C. Granger, Anal. Chem. 76, 1493–1499 (2004).
39. S. Plattner, R. Erb, J.P. Chervet, and H. Oberacher, Anal. Bioanal. Chem. 406, 213–224 (2014).
40. C. Zettersten, R. Lomoth, L. Hammarstrom, P.J.R. Sjoberg, and L. Nyholm, J. Electroanal. Chem. 590, 90–99 (2006).
41. C.F. Bokman, C. Zettersten, P.J.R. Sjoberg, and L. Nyholm, Anal. Chem. 76, 2017–2024 (2004).
42. H.T. Deng and G.J. Van Berkel, Electroanalysis 11, 857–865 (1999).
43. S.M. van Leeuwen, H. Hayen, and U. Karst, Anal. Bioanal. Chem. 378, 917–925 (2004).
44. H. Hayen and U. Karst, Anal. Chem. 75, 4833–4840 (2003).
45. H. Iwahashi, J. Chromatogr. B 736, 237–245 (1999).
46. H. Iwahashi and T. Ishii, J. Chromatogr. A 773, 23–31 (1997).
47. A. Baumann, W. Lohmann, B. Schubert, H. Oberacher, and U. Karst, J. Chromatogr. A 1216, 3192–3198 (2009).
48. E. Nouri-Nigjeh, H.P. Permentier, R. Bischoff, and A.P. Bruins, Analytical Chemistry 83, 5519–5525 (2011).
49. E. Nouri-Nigjeh, R. Bischoff, A.P. Bruins, and H.P. Permentier, Analyst 136, 5064–5067 (2011).
50. W. Lohmann and U. Karst, Anal. Chem. 79, 6831–6839 (2007).
51. U. Jurva, H.V. Wikstrom, and A.P. Bruins, Rapid Commun. Mass Spectrom. 16, 1934–1940 (2002).
52. T. Johansson, L. Weidolf, and U. Jurva, Rapid Commun. Mass Spectrom. 21, 2323–2331 (2007).
53. E. Nouri-Nigjeh, H.P. Permentier, R. Bischoff, and A.P. Bruins, Anal. Chem. 82, 7625–7633 (2010).
54. E. Nouri-Nigjeh, A.P. Bruins, R. Bischoff, and H.P. Permentier, Analyst 137, 4698–4702 (2012).
55. G. Hambitzer and J. Heitbaum, Anal. Chem. 58, 1067–1070 (1986).
56. G. Hambitzer and J. Heitbaum, J. Electrochem. Soc. 134, C496-C496 (1987).
57. K.J. Volk, R.A. Yost, and A. Brajter-Toth, Anal. Chem. 64, 21A–33A (1992).
58. K.J. Volk, M.S. Lee, R.A. Yost, and A. Brajter-Toth, Anal. Chem. 60, 720–722 (1988).
59. K. Volk, R. Yost, and A. Brajter-Toth, J. Electrochem. Soc. 134, C500–C500 (1987).
60. U. Jurva, H.V. Wikstrom, L. Weidolf, and A.P. Bruins, Rapid Commun. Mass Spectrom. 17, 800–810 (2003).
61. U. Jurva, H.V. Wikstrom, and A.P. Bruins, Rapid Commun. Mass Spectrom. 14, 529–533 (2000).
62. U. Bussy, Y.W. Chung-Davidson, K. Li, and W.M. Li, Anal. Bioanal. Chem. 406, 7253–7260 (2014).
63. K.G. Madsen, J. Olsen, C. Skonberg, S.H. Hansen, and U. Jurva, Chem. Res. Toxicol. 20, 821–831 (2007).
64. T. Johansson, U. Jurva, G. Gronberg, L. Weidolf, and C. Masimirembwa, Drug Metab. Dispos. 37, 571–579 (2009).
65. W. Lohmann and U. Karst, Anal. Bioanal. Chem. 386, 1701–1708 (2006).
66. K.G. Madsen, C. Skonberg, U. Jurva, C. Cornett, S.H. Hansen, T.N. Johansen, and J. Olsen, Chem. Res. Toxicol. 21, 1107–1119 (2008).
67. H. Faber, D. Melles, C. Brauckmann, C.A. Wehe, K. Wentker, and U. Karst, Anal. Bioanal. Chem. 403, 345–354 (2012).
68. T.J. Mali'n, L. Weidolf, N. Castagnoli, and U. Jurva, Rapid Commun. Mass Spectrom. 24, 1231–1240 (2010).
69. K.G. Madsen, G. Gronberg, C. Skonberg, U. Jurva, S.H. Hansen, and J. Olsen, Chem. Res. Toxicol. 21, 2035–2041 (2008).
70. K. Tahara, T. Nishikawa, Y. Hattori, S. Iijima, Y. Kouno, and Y. Abe, J. Pharm. Biomed. Anal. 50, 1030–1036 (2009).
71. K. Tahara, Y. Yano, K. Kanagawa, Y. Abe, J. Yamada, S. Iijima, M. Mochizuki, and T. Nishikawa, Chem. Pharm. Bull. 55, 1207–1212 (2007).
72. M. Lu, C. Wolff, W.D. Cui, and H. Chen, Anal. Bioanal. Chem. 403, 355–365 (2012).
73. K. Nozaki, I. Osaka, H. Kawasaki, and R. Arakawa, Anal. Sci. 25, 1197–1201 (2009).
74. K. Nozaki, H. Kitagawa, S. Kimura, A. Kagayama, and R. Arakawa, J. Mass Spectrom. 41, 606–612 (2006).
75. U. Bussy, M. Delaforge, C. El-Bekkali, V. Ferchaud-Roucher, M. Krempf, I. Tea, N. Galland, D. Jacquemin, and M. Boujtita, Anal. Bioanal. Chem. 405, 6077–6085 (2013).
76. U. Bussy, I. Tea, V. Ferchaud-Roucher, M. Krempf, V. Silvestre, N. Galland, D. Jacquemin, M. Andresen-Bergstrom, U. Jurva, and M. Boujtita, Anal. Chim. Acta 762, 39–46 (2013).
77. R.G. de Lima, P.S. Bonato, and R.S. da Silva, J. Pharm. Biomed. Anal. 32, 337–343 (2003).
78. S. Jahn, A. Baumann, J. Roscher, K. Hense, R. Zazzeroni, and U. Karst, J. Chromatogr. A 1218, 9210–9220 (2011).
79. S. Jahn, B. Seiwert, S. Kretzing, G. Abraham, R. Regenthal, and U. Karst, Anal. Chim. Acta 756, 60–72 (2012).
80. T.A. Getek, W.A. Korfmacher, T.A. Mcrae, and J.A. Hinson, J. Chromatogr. 474, 245–256 (1989).
81. S. Jahn, W. Lohmann, S. Bomke, A. Baumann, and U. Karst, Anal. Bioanal. Chem. 402, 461–471 (2012).
82. U. Jurva, A. Holmen, G. Gronberg, C. Masimirembwa, and L. Weidolf, Chem. Res. Toxicol. 21, 928–935 (2008).
83. W. Lohmann, H. Hayen, and U. Karst, Anal. Chem. 80, 9714–9719 (2008).
84. S. Jahn, H. Faber, R. Zazzeroni, and U. Karst, Rapid Commun. Mass Spectrom. 26, 1453–1464 (2012).
85. S. Jahn, H. Faber, R. Zazzeroni, and U. Karst, Rapid Commun. Mass Spectrom. 26, 1415–1425 (2012).
86. D. Melles, T. Vielhaber, A. Baumann, R. Zazzeroni, and U. Karst, J. Chromatogr. B 913, 106–112 (2013).
87. L. Chen, S. Kuppers, Z. Wang, X.Y. Xiang, and S.W. Cao, Environ. Chem. Lett. 12, 329–334 (2014).
88. L. Chen, D. Hofmann, E. Klumpp, X.Y. Xiang, Y.X. Chen, and S. Kuppers, Chemosphere 89, 1376–1383 (2012).
89. T. Hoffmann, D. Hofmann, E. Klumpp, and S. Kuppers, Anal. Bioanal. Chem. 399, 1859–1868 (2011).
90. H. Faber, H. Lutze, P.L. Lareo, L. Frensemeier, M. Vogel, T.C. Schmidt, and U. Karst, J. Chromatogr. A 1343, 152–159 (2014).
91. W. Lohmann, R. Dotzer, G. Gutter, S.M. Van Leeuwen, and U. Karst, J. Am. Soc. Mass Spectrom. 20, 138–145 (2009).
92. S. Jahn, S. Beuck, I. Moller, M. Thevis, and U. Karst, Anal. Methods 5, 1214–1224 (2013).
93. M. Thevis, W. Lohmann, Y. Schrader, M. Kohler, W. Bornatsch, U. Karst, and W. Schanzer, Eur. J. Mass Spectrom. 14, 163–170 (2008).
94. M. Smoluch, P. Mielczarek, E. Reszke, G.M. Hieftje, and J. Silberring, Analyst 139, 4350–4355 (2014).
95. P. Mielczarek, H. Raoof, J.H. Kotlinska, P. Stefanowicz, Z. Szewczuk, P. Suder, and J. Silberring, Eur. J. Mass Spectrom. 20, 279–285 (2014).
96. A.J. Pedersen, L. Ambach, S. Konig, and W. Weinmann, Bioanalysis 6, 2607–2621 (2014).
97. P.H. Gamache, D.F. Meyer, M.C. Granger, and I.N. Acworth, J. Am. Soc. Mass Spectrom. 15, 1717–1726 (2004).
98. N.A. Mautjana, D.W. Looi, J.R. Eyler, and A. Brajter-Toth, Electroanalysis 22, 79–89 (2010).
99. N.A. Mautjana, D.W. Looi, J.R. Eyler, and A. Brajter-Toth, Electrochim. Acta 55, 52–58 (2009).
100. N.A. Mautjana, J. Estes, J.R. Eyler, and A. Brajter-Toth, Electroanalysis 20, 2501–2508 (2008).
101. H. Oberacher, B. Schubert, K. Libiseller, and A. Schweissgut, Anal. Chim. Acta 770, 121–131 (2013).
102. H. Oberacher, M. Pavlic, K. Libiseller, B. Schubert, M. Sulyok, R. Schuhmacher, E. Csaszar, and H.C. Kofeler, J. Mass Spectrom. 44, 485–493 (2009).
103. H.P. Permentier, U. Jurva, B. Barroso, and A.P. Bruins, Rapid Commun. Mass Spectrom. 17, 1585–1592 (2003).
104. C.S. McClintock and R.L. Hettich, Anal. Chem. 85, 213–219 (2013).
105. C. McClintock, V. Kertesz, and R.L. Hettich, Anal. Chem. 80, 3304–3317 (2008).
106. J. Roeser, H.P. Permentier, A.P. Bruins, and R. Bischoff, Anal. Chem. 82, 7556–7565 (2010).
107. J. Roeser, N.F.A. Alting, H.P. Permentier, A.P. Bruins, and R. Bischoff, Anal. Chem. 85, 6626–6632 (2013).
108. Y. Zhang, W.D. Cui, H. Zhang, H.D. Dewald, and H. Chen, Anal. Chem. 84, 3838–3842 (2012).
109. Y. Zhang, H.D. Dewald, and H. Chen, J. Proteome Res. 10, 1293–1304 (2011).
110. Y. Zhang, Z.Q. Yuan, H.D. Dewald, and H. Chen, Chem. Commun. 47, 4171–4173 (2011).
111. S. Nicolardi, M. Giera, P. Kooijman, A. Kraj, J.P. Chervet, A.M. Deelder, and Y.E.M. van der Burgt, J. Am. Soc. Mass Spectrom. 24, 1980–1987 (2013).
112. A. Kraj, H.J. Brouwer, N. Reinhoud, and J.P. Chervet, Anal. Bioanal. Chem. 405, 9311–9320 (2013).
113. S. Nicolardi, A.M. Deelder, M. Palmblad, and Y.E.M. van der Burgt, Anal. Chem. 86, 5376–5382 (2014).
114. S. Mysling, R. Salbo, M. Ploug, and T.J.D. Jorgensen, Anal. Chem. 86, 340–345 (2014).
115. T.C. Rohner, J.S. Rossier, and H.H. Girault, Electrochem. Commun. 4, 695–700 (2002).
116. T.C. Rohner and H.H. Girault, Rapid Commun. Mass Spectrom. 19, 1183–1190 (2005).
117. L. Dayon, C. Roussel, and H.H. Girault, J. Proteome Res. 5, 793–800 (2006).
118. M. Prudent, J.S. Rossier, N. Lion, and H.H. Girault, Anal. Chem. 80, 2531–2538 (2008).
119. Y. Lu, M. Prudent, L.A. Qiao, M.A. Mendez, and H.H. Girault, Metallomics 2, 474–479 (2010).
120. E. Palecek, Nature 188, 656-657 (1960).
121. P. Subramanian and G. Dryhurst, J. Electroanal. Chem. 224, 137–162 (1987).
122. G. Dryhurst and G.F. Pace, J. Electrochem. Soc. 117, 1259–1264 (1970).
123. R.N. Goyal, S.M. Sondhi, and A.M. Lahoti, New J. Chem. 29, 587–595 (2005).
124. F. Pitterl, J.P. Chervet, and H. Oberacher, Anal. Bioanal. Chem. 397, 1203–1215 (2010).
125. S. Plattner, R. Erb, F. Pitterl, H.J. Brouwer, and H. Oberacher, J. Chromatogr. B 883, 198–204 (2012).
126. H. Oberacher, R. Erb, S. Plattner, and J.P. Chervet, Trends Anal. Chem. , in press.
127. A. Baumann, W. Lohmann, S. Jahn, and U. Karst, Electroanalysis 22, 286–292 (2010).
128. R. Scholz, P. Palatzky, and F.M. Matysik, Anal. Bioanal. Chem. 406, 687–694 (2014).
129. M. Cindric, M. Vojs, and F.-M. Matysik, Electroanalysis 27, 234–241 (2015).
130. R. Singh and P.B. Farmer, Carcinogenesis 27, 178–196 (2006).
131. K.M. Li, J. Byun, M.L. Gross, D. Zamzow, R. Jankowiak, E.G. Rogan, and E.L. Cavalieri, Chem. Res. Toxicol. 12, 749–757 (1999).
132. D.W. Looi, J.R. Eyler, and A. Brajter-Toth, Electrochim. Acta 56, 2633–2640 (2011).
133. R.J. Klose and A.P. Bird, Trends in Biochem. Sci. 31, 89–97 (2006).
134. S. Kriaucionis and N. Heintz, Science 324, 929–930 (2009).
135. M. Tahiliani, K.P. Koh, Y. Shen, W.A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L.M. Iyer, D.R. Liu, L. Aravind, and A. Rao, Science 324, 930–935 (2009).
136. S. Ito, L. Shen, Q. Dai, S.C. Wu, L.B. Collins, J.A. Swenberg, C. He, and Y. Zhang, Science 333, 1300–1303 (2011).
137. Y.F. He, B.Z. Li, Z. Li, P. Liu, Y. Wang, Q.Y. Tang, J.P. Ding, Y.Y. Jia, Z.C. Chen, L. Li, Y. Sun, X.X. Li, Q. Dai, C.X. Song, K.L. Zhang, C. He, and G.L. Xu, Science 333, 1303–1307 (2011).
138. D. Cortazar, C. Kunz, J. Selfridge, T. Lettieri, Y. Saito, E. MacDougall, A. Wirz, D. Schuermann, A.L. Jacobs, F. Siegrist, R. Steinacher, J. Jiricny, A. Bird, and P. Schar, Nature 470, 419–U210 (2011).
139. K.P. Koh and A. Rao, Current Opinion in Cell Biology 25, 152–161 (2013).
140. C.X. Song and C. He, Trends in Biochem. Sci. 38, 480–484 (2013).
141. C.G. Spruijt, F. Gnerlich, A.H. Smits, T. Pfaffeneder, P.W.T.C. Jansen, C. Bauer, M. Munzel, M. Wagner, M. Muller, F. Khan, H.C. Eberl, A. Mensinga, A.B. Brinkman, K. Lephikov, U. Muller, J. Walter, R. Boelens, H. van Ingen, H. Leonhardt, T. Carell, and M. Vermeulen, Cell 152, 1146–1159 (2013).
142. M. Mellen, P. Ayata, S. Dewell, S. Kriaucionis, and N. Heintz, Cell 151, 1417–1430 (2012).
143. J.R. Wagner and J. Cadet, Acc. Chem. Res. 43, 564–571 (2010).
144. J. Cadet and J.R. Wagner, Mutat. Res. 764, 18–35 (2014).
145. A. Grand, C. Morell, V. Labet, J. Cadet, and L.A. Eriksson, J. Phys. Chem. A 111, 8968–8972 (2007).

New TRC Facility Accelerates Innovation and Delivery
April 25th 2025We’ve expanded our capabilities with a state-of-the-art, 200,000 sq ft TRC facility in Toronto, completed in 2024 and staffed by over 100 PhD- and MSc-level scientists. This investment enables the development of more innovative compounds, a broader catalogue and custom offering, and streamlined operations for faster delivery. • Our extensive range of over 100,000 high-quality research chemicals—including APIs, metabolites, and impurities in both native and stable isotope-labelled forms—provides essential tools for uncovering molecular disease mechanisms and exploring new opportunities for therapeutic intervention.
New Guide: Characterising Impurity Standards – What Defines “Good Enough?”
April 25th 2025Impurity reference standards (IRSs) are essential for accurately identifying and quantifying impurities in pharmaceutical development and manufacturing. Yet, with limited regulatory guidance on how much characterisation is truly required for different applications, selecting the right standard can be challenging. To help, LGC has developed a new interactive multimedia guide, packed with expert insights to support your decision-making and give you greater confidence when choosing the right IRS for your specific needs.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)









