mRNA Characterization, from 5’ Cap to Poly (A): What Ion-Pair Reversed-Phase Liquid Chromatography (IP-RPLC) Analysis Can Tell You
Messenger RNA (mRNA) is a rapidly growing class of therapeutic compounds being studied to support vaccine development after its utilization during the COVID-19 pandemic. The manufacturing of mRNA consists of in vitro transcription reactions resulting in a product over 1000 nucleotides in length with a 5’Cap and a poly(A) tail. The 5’ Cap and the poly(A) tail help to stabilize the RNA molecule and improve its translation. To ensure safety and efficacy of mRNA vaccines, contaminants such as degradation products, incomplete capping, and shorter than expected poly(A) tails need to be characterized. This article will discuss an ion-pair reversed-phase liquid chromatography mass spectrometry (IP-RPLC–MS) workflow for the characterization of three mRNA key critical quality attributes: 5’ Cap, ORF, and poly(A) tail.
The introduction of messenger RNA (mRNA) therapeutics during the development of COVID-19 vaccines (1) has opened the door to applying this technology in other pharmaceutical and biotechnological areas. In recent years, we have witnessed exploring the capabilities of mRNA to treat various disorders such as genetic diseases (2) or cancer (3). The significant attention in recent years to mRNA therapeutics expanded the market size to $11.75 billion in 2023, with a forecast to grow at a CAGR of 17.05% from 2024 to 2030 (4).
Consequently, there has been an increased demand for mRNA manufacturing in recent years to meet global research and healthcare demands (5). While scaling up the production of mRNA, the industry must ensure that these meet all the critical quality attributes (CQAs) necessary for regulatory approval and clinical efficacy. The product must also be produced according to good manufacturing practices (GMP).
The mRNA therapeutics usually consist of complex structures of over 1000 nucleotides in length, including a 5’ cap, open reading frame (ORF), and a poly(A) tail. Any modification or irregularities that lead to additional degradation products, incomplete capping, or variation in the length of the poly(A) tails can significantly impact the safety and therapeutic effectiveness of mRNA products (6). This highlights the need to explore comprehensive analytical capabilities that confirm the integrity and functionality of the entire complex from a 5' cap to a poly(A) tail.
Among various techniques, ion-pair reversed-phase liquid chromatography (IP-RPLC) is becoming particularly promising because it combines the high resolving power of LC with the sensitivity and specificity of mass spectrometry (MS). IP-RPLC allows for detailed analysis of the complete mRNA sequence. The ion-pairing agents neutralize the charges on the mRNA, which enhances the retention and resolution of the sample. It allows for the recognition between different types of caps, such as Cap 0, Cap 1, and Cap 2, which are responsible for translational efficiency. The technique can also qualitatively measure the concentrations of the product and impurities within the sample. Implementing IP-RPLC within the workflows can enhance analytical capacities, resulting in better control of the manufacturing process and ultimately leading to safer and more effective mRNA final products.
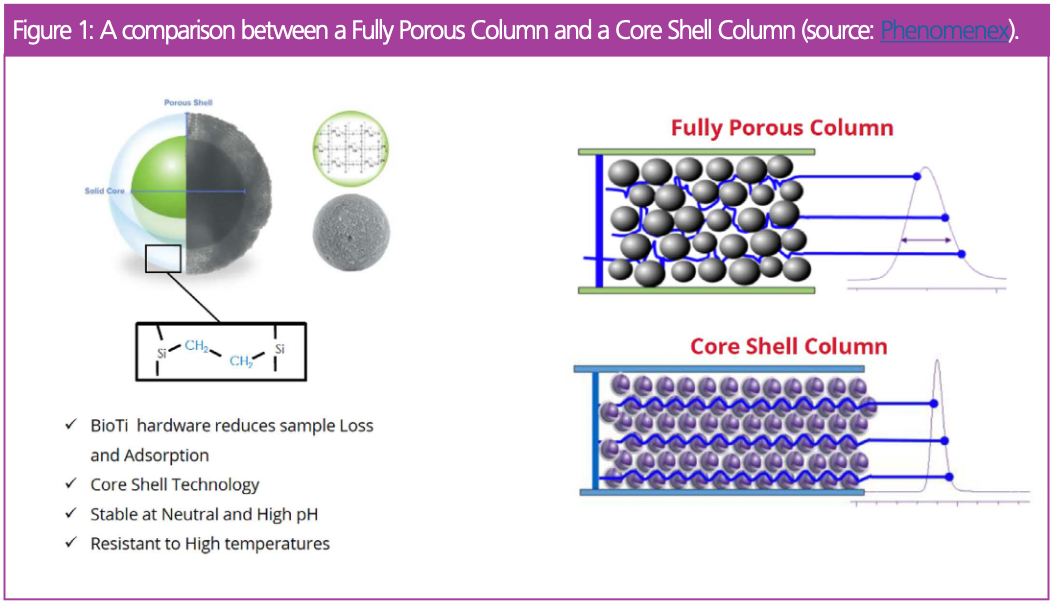
Experimental
In this study, the IP-RPLC workflow for mRNA characterization consisted of sample preparation through enzymatic digestion, LC–MS data collection of digested oligonucleotides, and bioinformatics post-analysis. A 10 μg of the mRNA sample, previously denatured with 1M urea at 90°C, was used for digestion. The mRNA was digested into oligonucleotides through enzymatic digestion with human RNase4 conducted for one hour to promote cleavage at U-A and U-G sites. The samples were analyzed using IP-RPLC and a Phenomenex Biozen Oligo column (Phenomonex).
Following separation, the eluted fractions were analyzed using a Sciex 7600 ZenoTOF mass spectrometer (Sciex). Post-acquisition, the mass spectra data underwent bioinformatics analysis using Protein Metrics software (Dotmatics).
Results and Discussion
Proper sample preparation is a crucial step for effective IP-RPLC analysis. In this study, the mRNA was digested using hRNase4, which effectively cleaves at U-A and U-G sites, producing oligonucleotides of optimal length for subsequent chromatographic separation. The use of T4 Polynucleotide Kinase (T4 PNK) facilitates the removal of the phosphate at the 3’ termini, leaving fully hydroxylated oligonucleotides. The appropriate size, as 6-mers to 20-mers, is particularly important for both sequence separation with LC and identification with MS compared to longer fragments. The latter usually do not fragment well during MS2 and pose ionization challenges, making them less suitable for effective analysis.
The column used minimizes non-specific adsorption to the column. This is particularly relevant when handling precious samples available in small amounts, such as mRNA. This column is also robust under various pH levels or elevated temperatures.
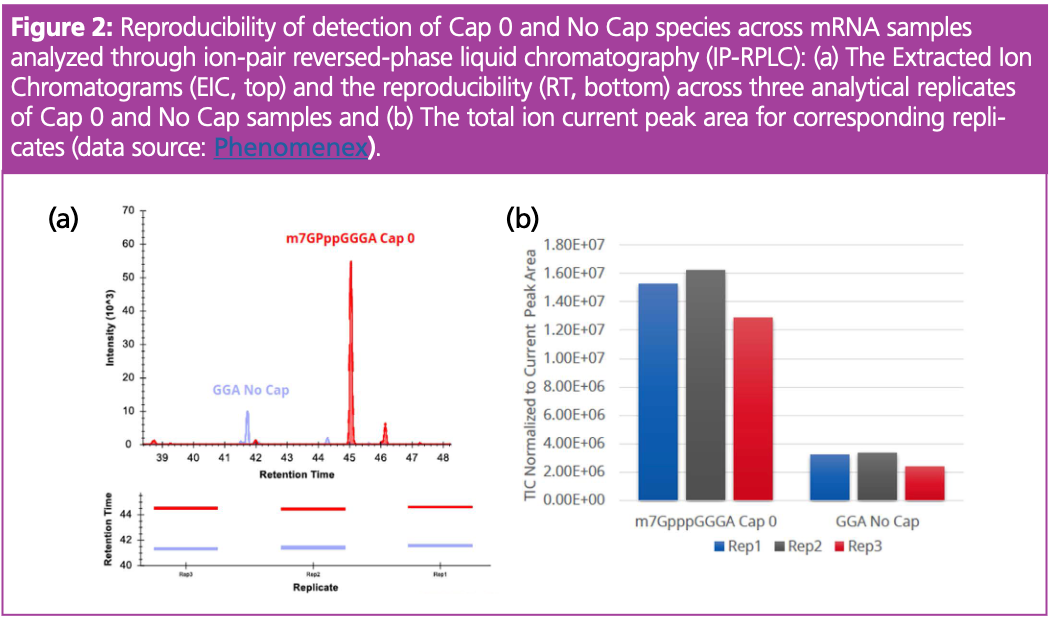
Our results demonstrated the effectiveness of using IP-RPLC for mRNA 5’ cap characterization. Figure 2(a) shows a chromatogram with peaks corresponding to the m7GpppGGGA (Cap 0) and GGA (No Cap), while Figure 2(b) presents normalized peak areas. Overall, the technique consistently identified variants of Cap 0 and No Cap species with small variability across replicates. For instance, the analysis of the replicates of Cap 0 (m7GpppGGGA) spectra with intensities of 1.6E ± 07, 1.4E ± 07, and 1.5E ± 07 counts resulted in a standard deviation of 0.15. Similarly, a small standard deviation was observed for the measurement of No Cap samples. This highlights the capability of the technique to deliver repeatable and accurate measurements essential for qualitative and quantitative assessments of caps, which are crucial for the translation and stability of mRNA.
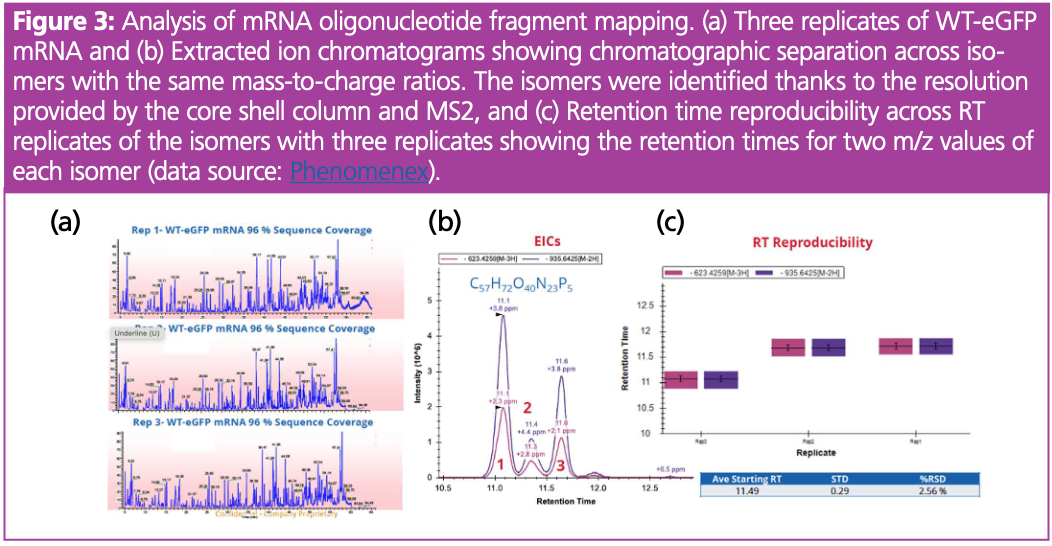
We further employed the analytical techniques for mRNA oligo fragment mapping. The total ion chromatogram (TIC) represented the peaks corresponding to various oligonucleotide fragments (Figure 2[a]). Importantly, the bioinformatics analysis showed that the mRNA oligonucleotide mapping achieved a sequence coverage of 96%. This high sequence coverage is essential because it ensures that most of the mRNA sequence is accounted for, which is critical for reliable interpretations in therapeutic mRNA applications.
Importantly, we showed that total ion chromatograms (TIC) were consistent between sample replicates, showing similar peak shapes and retention times, which highlights the analytical reproducibility of this approach. Moreover, the chromatographic analysis resulted in the clear separation of peaks, allowing for the identification of three different isomers. This identification was based on both their chromatographic resolution and MS2 fragmentation because the mass-to-charge ratios (623.4259[M-3H] and 935.6425[M-2H]) are the same for these isomers (Figure 3[a] and 3[c]). This shows the importance of good chromatographic practices which allow for accurate and detailed spectroscopic analysis. Overall, high-resolution mass spectrometry enables the differentiation of oligo isomers, highlighting that the technique is crucial for understanding mRNA modification and structural variability.
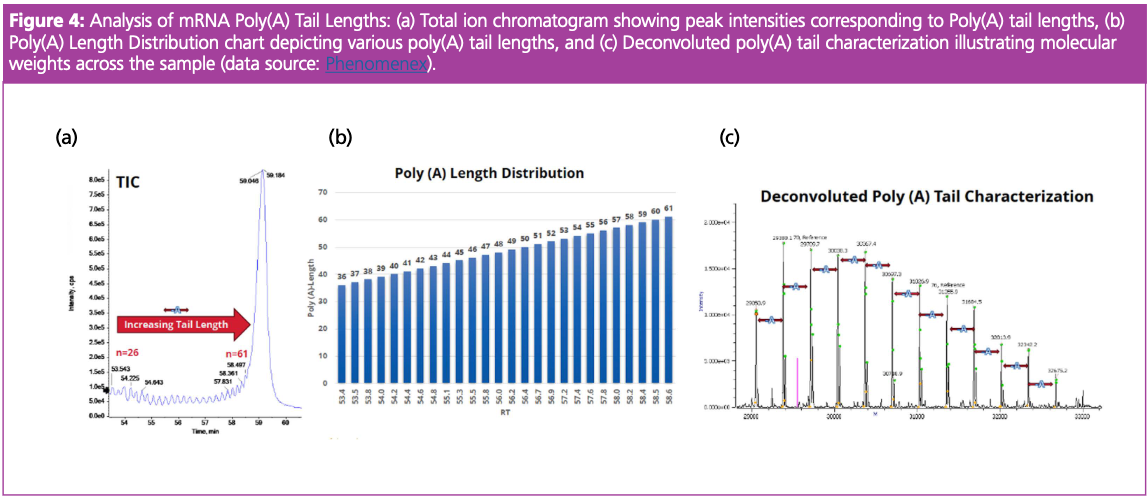
Ion-pairing RPLC coupled with MS using MS1 was further applied to assess the distribution and heterogeneity of mRNA 3’ poly(A) Tail Length. The TIC displayed the peaks representing different mRNA polyadenylation lengths, indicating that the sample contained a variety of poly(A) tail lengths (Figure 4[a]). The accompanying poly(A) Tail length distribution chart represented the chromatographic resolution observed across the poly(A) tail lengths, with the y-axis indicating retention time (Figure 4[b]). The deconvoluted mass spectra of the mRNA sample, identified using accurate mass, showed the loss of 329 daltons, which is related to the loss of adenine residues from the poly(A) tail indicating the presence of shorter poly(A) tails due to cleavage or incomplete synthesis (Figure 4[c]). The technique resolved each mass peak, allowing for annotation of exact mass values, demonstrating the high resolution of the mass spectrometry technique and confirming its utility for analysis of the distribution and heterogeneity of mRNA 3’poly (A) Tail. Shorter tails usually lead to faster mRNA degradation, while longer tails can provide additional stability and efficiency during translation. Overall, IP-RPLC can be a robust analytical tool for mRNA, contributing to the development of functional mRNA-based therapeutics.
One of the additional challenges associated with the IP-RPLC analysis is generating a high volume of data. the software confirmed the identity and integrity of mRNA constructs by applying algorithms for deconvolution against known samples (oligonucleotide spectrum matching). Applying bioinformatics platforms also enables the identification of post-transcriptional modifications within the sample and degradation products, providing a holistic view of the composition and biological efficacy of mRNA-based therapeutics. Overall, IP-RPLC and bioinformatics analysis provided a comprehensive analysis of mRNA molecules.
Conclusion
The current market analysis foresees the expansion of mRNA technology to various biotechnological and pharmacological fields, underlining the importance of robust analytical techniques that will allow for quality checks and the delivery of safe and efficient products. This work presented a unified workflow integrating the appropriate sample digestion, IP-RPLC, and advanced bioinformatics analysis.
The workflow enabled the characterization of the mRNA 5’ cap, mRNA oligo fragment, and mRNA 3’ poly (A) tail length, which are crucial for maintaining the stability and translation efficiency of mRNA-based products. Overall, this workflow demonstrated high-resolution, accuracy, and reproducibility across the samples, presenting a holistic approach to mRNA analysis. Advanced analytical platforms such as IP-RPLC can help develop mRNA for various therapeutic applications, streamlining biotechnological innovation.
References
(1) Pardi, N.; Hogan, M. J.; Weissman, D. Recent Advances in MRNA Vaccine Technology. Curr. Opin. Immunol. 2020, 65, 14–20. DOI: 10.1016/j.coi.2020.01.008
(2) Shen, G.; Liu, J.; Yang, H.; Xie, N.; Yang, Y. MRNA Therapies: Pioneering a New Era in Rare Genetic Disease Treatment. J. Control. Release 2024, 369, 696–721. DOI: 10.1016/j.jconrel.2024.03.056
(3) Liu, C.; Shi, Q.; Huang, X.; Koo, S.; Kong, N.; Tao, W. MRNA-Based Cancer Therapeutics. Nat. Rev. Cancer 2023, 1–18. DOI: 10.1038/s41568-023-00586-2
(4) mRNA Therapeutics Market Size & Share Report, 2022-2030. Grandview Research. https://www.grandviewresearch.com/industry-analysis/mrna-therapeutics-market-report
(5) Youssef, M.; Hitti, C.; Puppin Chaves Fulber, J.; Kamen, A. A. Enabling MRNA Therapeutics: Current Landscape and Challenges in Manufacturing. Biomolecules 2023, 13 (10), 1497. DOI: 10.3390/biom13101497
(6) Qin, S.; Tang, X.; Chen, Y.; Chen, K.; Fan, N.; Xiao, W.; Zheng, Q.; Li, G.; Teng, Y.; Wu, M.; Song, X. MRNA-Based Therapeutics: Powerful and Versatile Tools to Combat Diseases. Signal Transduct. Target. Ther. 2022, 7 (1), 1–35. DOI: 10.1038/s41392-022-01007-w
About the Author
Roxana Eggleston-Rangel joined Phenomenex in 2018 as an Applications Scientist and is currently working with the applications team to produce analytical solutions for scientist in the biopharma industry. Roxana’s current work focuses on characterization of mRNA, sgRNA and siRNA.
Prior to Phenomenex, Roxana worked for Caltech and the University of Southern California developing LC–MS workflows for the proteomics community.
Roxana received her bachelor’s and master’s degree in Chemistry and Biochemistry from California State University, Los Angeles. Direct correspondence to: RoxanaE@phenomenex.com

New Method Explored for the Detection of CECs in Crops Irrigated with Contaminated Water
April 30th 2025This new study presents a validated QuEChERS–LC-MS/MS method for detecting eight persistent, mobile, and toxic substances in escarole, tomatoes, and tomato leaves irrigated with contaminated water.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)












