Membrane Proteins
The Application Notebook
Membrane proteins - together with lipids - make up biological membranes that are essential for life. In order to understand the role of membrane proteins in assisting membranes to carry out many different functions, it is of great importance to understand the structure of those proteins.
Membrane proteins - together with lipids - make up biological membranes that are essential for life. In order to understand the role of membrane proteins in assisting membranes to carry out many different functions, it is of great importance to understand the structure of those proteins.
Membrane protein is generally soluble only in the presence of micelles; thus, it is very difficult to characterize the oligomerization state of the membrane protein in a lipid-containing solvent. In this application note we demonstrate the use of multi-angle light scattering (MALS) detection in combination with UV absorption and differential refractive index (DRI) detection to determine the molar masses (MM) of both the core protein and the entire protein-lipid complex.
The chromatograms of one membrane protein were obtained from size-exclusion chromatography (SEC). SEC can often refer to fast protein liquid chromatography, or, FPLC (Amersham Biosciences, Uppsala, Sweden). In this experimental set-up, a DAWN MALS detector was coupled to a UV (280 nm) and DRI detector, and the resulting traces are shown in Figure 1.
Figure 1: Chromatograms of a membrane protein obtained from a LS (top), DRI (middle), and UV at 280 nm (bottom) detectors.
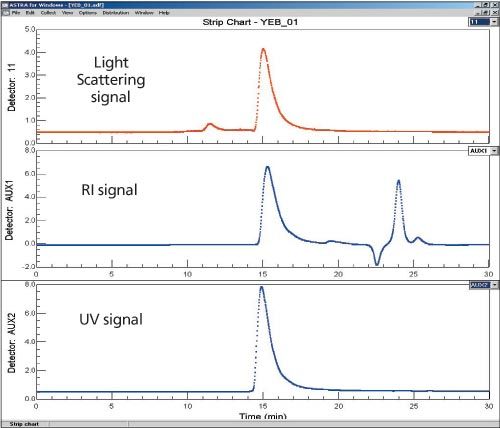
In order to keep the membrane protein in solution, it was necessary to use a mobile phase that contained lipids at greater than the critical micelle concentration. Since the membrane protein-lipid complex has quite a different conformation and probably different adsorption characteristics to the column packing than globular standard proteins, the elution property of membrane proteins and globular proteins are very different. As a result, the traditional column calibration method fails to provide any estimation on molar mass using elution time.
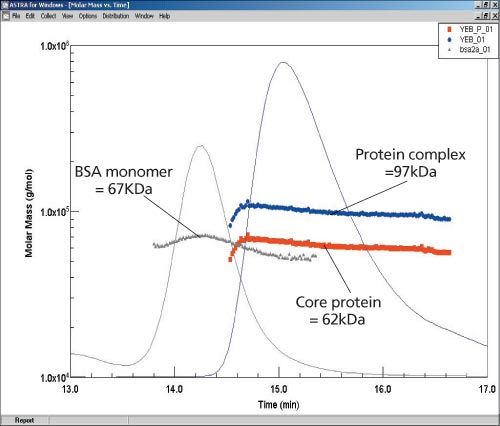
ASTRA software’s protein conjugate algorithm analyses the data from the MALS, UV, and DRI detectors to determine molar masses of the core membrane protein, lipid micelle, and protein-lipid complex, as seen in Figure 2. These data suggest that the membrane protein is in a monomeric state (62 kDa).
Figure 2: The analysis based on data from LS, UV, and DRI detectors reveals molar masses for the core protein and protein-lipid complex are 62 and 97 kD, respectively. The results from BSA demonstrate that the SEC properties of these two protein samples are very different.
This example demonstrates clearly that the combination of MALS, UV, and DRI detection is an unique and powerful tool in characterizing membrane proteins in particular, and other modified proteins - such as pegylated and glycosylated proteins - in general.

Wyatt Technology Corporation
630 Hollister Avenue, Santa Barbara, California 93117, USA
Tel: (805) 681 9009 Fax: (805) 681 0123
E-mail: info@wyatt.com Website: www.wyatt.com


.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)










