Intact Mass Analysis–Based Multi-Attribute Methods (iMAMs) for Characterization of Biopharmaceuticals
Multi-attribute methods (MAMs) are becoming increasingly popular for their ability to analyze multiple critical quality attributes (CQA) in a single workflow. This capability becomes particularly attractive for a product class such as monoclonal antibodies, which are large and complex, and have many CQAs that need to be monitored and controlled during their manufacturing so as to deliver consistent product quality. In an earlier installment, we discussed the role of liquid chromatography and mass spectrometry in MAMs. In this article, we focus on intact mass analysis–based multi-attribute methods (iMAMs), a suitable alternative that can complement standard MAMs or be used when there is a need for rapid turnaround and monitoring of only a limited number of CQAs. Multiple case studies are presented to elucidate this concept.
Monoclonal antibodies (mAbs) are the fastest-growing class of therapeutics used in the treatment of wide variety of diseases. The complex nature of mAbs demands characterization by multiple advanced analytical tools that can deliver efficient and enhanced monitoring of the numerous critical quality attributes (CQAs) that a typical mAb product has. Traditionally, chromatographic methods such as ion-exchange or reversed-phase chromatography or capillary or imaged capillary isoelectric focusing separation methods are used for product characterization and for quality control (QC) testing. However, these techniques can neither distinguish between species varying with respect to site-specific modifications nor identify these modifications (1). Advancement in mass spectrometry (MS), especially in terms of resolution, robustness, and ease of use, have made MS the tool of choice for both product characterization as well as for identification of site-specific modifications.
The original multi-attribute method (MAM) was proposed as a means of monitoring multiple site-specific CQAs using an ultrahigh-pressure
liquid chromatography–mass spectrometry (UHPLC–MS) method based on a peptide mapping tool (2,3). istics such as purity, size, and charge. MAMs have undergone development from the conventional peptide map method for characterization of monoclonal antibodies to a two-part method comprising targeted monitoring and non-targeted new peak detection (NPD) (2). The topics of targeted monitoring and NPD, crucial components of MAMs, have been elaborated in a previous article on this topic (4). That article discussed critical experimental parameters necessary for employing an NPD approach in MAMs to ensure accurate results while eliminating false negatives. In addition to peptide-map based MAMs, complementary subunit or intact mass–based MAMs have been recently introduced to monitor various attributes in a single LC–MS method such as identification of glycosylation and oxidation to achieve faster analysis and simpler sample preparation (5,6). In this article, we describe intact mass analysis–based MAMs (iMAMs) and their application in product characterization.
Advantages of MAMs
Traditional analytical techniques evaluate method-specific product characteristics such as purity, size, and charge. These techniques are time-consuming, resource-intensive, and have a limited ability to identify site-specific modifications in product quality (7). When compared to traditional approaches, MAMs offer residue-specific identification, quantitative monitoring, and increased understanding of post-translational modifications (PTMs) and chemical modifications in the product. Some key advantages that MAMs offer include improved detection sensitivity, accuracy, efficiency, and risk assessment (8). Additionally, a MAM can be carried out in a single run, which reduces complexity, cost, and time compared to traditional analytical methods. MAMs can also detect low-abundant modifications in the mAbs, which are difficult to detect with conventional methods, making them more effective in monitoring product quality and making it easier to identify any process- or product-related changes (9). Furthermore, MAMs can be applied to multiple batches of a product in a single run, thereby offering better coverage for CQAs. MAMs can streamline the life cycle management processes by easing regulatory barriers because a single method is used in place of numerous conventional methods. Using MAMs during the development and production of mAbs can help ensure regulatory compliance and speed up the approval of new products. Regulatory organizations, like the US Food and Drug Administration (FDA) and the Euro- pean Medicines Agency (EMA), are beginning to recognize MAMs as a potent tool for evaluating product quality (10).
Applications for Product Characterization
MAMs can be applied at the top-down or bottom-up level for monitoring and characterization of product quality during various stages of production from cell line development to the final product. Critical quality monitoring at an early stage minimizes the impact on product quality and prevents potential recalls. Generally, a MAM based on a bottom-up approach employed for in-depth characterization of a product can provide a comprehensive examination of the residue sequence and sites of PTMs. However, this approach requires extensive sample preparation, which, despite major advancements in automation, continues to be prone to artifacts such as induced modifications and a lack of reproducibility (11). On the other hand, a subunit or intact-mass analysis requires minimum sample preparation and can monitor multiple attributes—such as purity, size variants, glycosylation and charge variants—in a single LC–MS method (12–14). This approach can provide an enhanced control strategy for reporting attributes, such as titer, presence of dimer or fragments, or antibody–drug conjugations, something that would not be possible to monitor through a standard MAM. This information can be critical for lot release and stability, as well as for setting biologically relevant specifications. Moreover, MAMs can play a crucial role by monitoring attributes and incorporating biologically relevant information for characterization of CQAs during the process development phase and facilitating QC analysis for product release. The data collected during product development can assist in the determination and validation of the CQAs to be monitored during product release and stability testing, as well as determining their acceptance criteria. Additionally, MAMs can be employed as a process analytical technology (PAT) tool to continuously evaluate CQAs. This makes it possible to spot process deviations early on and ensures consistency in product quality. Historical data for specific quality attributes can be obtained by reanalyzing data from prior runs, batches, and stability samples if new knowledge about the product is gained or if novel impurities or product variants are discovered. This feature is not available for most conventional QC methods because of their lack of specificity.
This ability of MAMs has the potential to revolutionize the approach for establishing product control strategies.
Applications for Biosimilarity
A growing number of biosimilars are receiving approval by regulatory agencies as the patents on the existing mAb products expire. Establishing analytical and functional biosimilarity serves as the foundation for biosimilar development programs. MAMs have gained immense popularity because of their ability to monitor multiple attributes across a large number of samples with higher accuracy than conventional approaches. Conventional methods for assessing biosimilars typically involve comparing the biosimilar to the reference (innovator) product using a variety of analytical methods. However, 1D approaches offer limited characterization capability. For example, traditional ion-exchange chromatography can differentiate charge variants in terms of acidic and basic variants, but is unable to identify the modifications that are present in the variants. On the other hand, iMAMs or standard MAMs offer enhanced capability toward characterization of the variants (Table I). MAMs can identify minor differences between a biosimilar and reference product that may otherwise not be identified by conventional methods.
Case Studies
MAMs are useful when residue-specific monitoring is not required, and when rapid analysis is desired for monitoring attributes such as aggregates, dimers or fragments, or charge variants. A recently published article showed a MAM workflow to monitor charge heterogeneity of mAbs, described here as a case study (14).
Case Study I: Monitoring Charge Based Heterogeneity
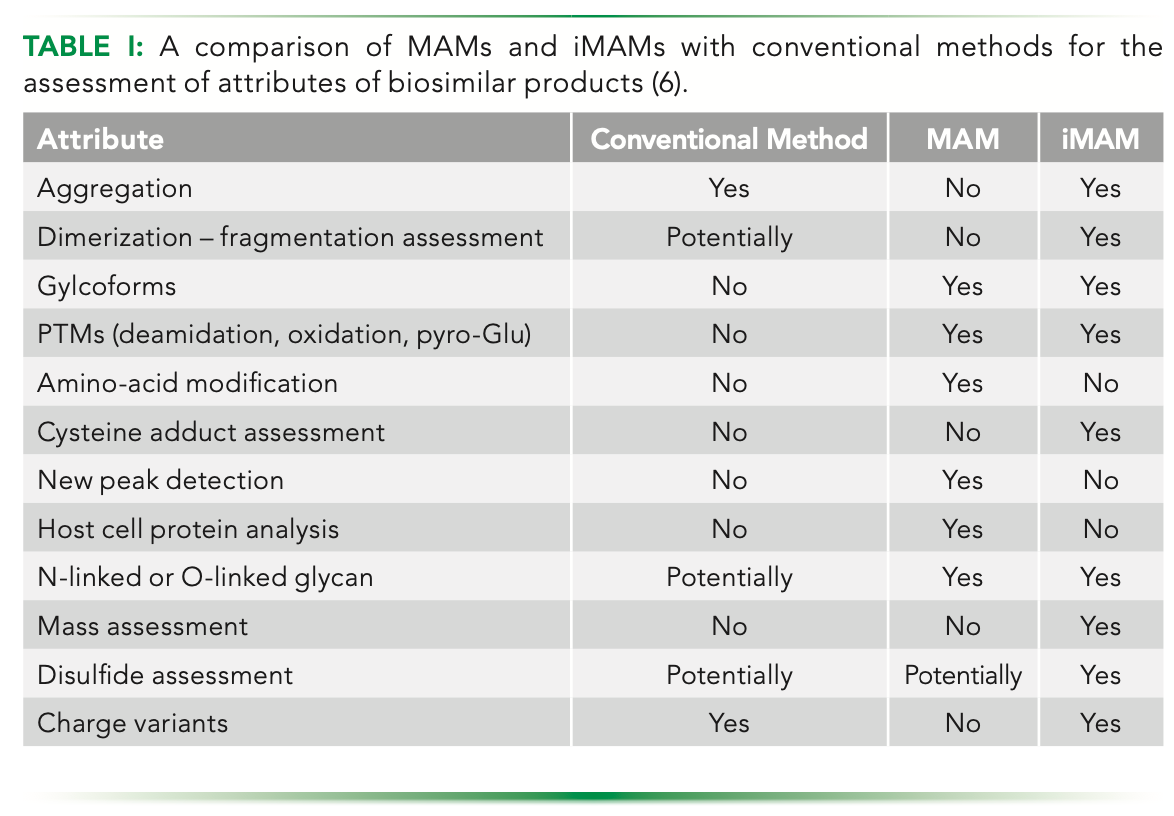
Two-dimensional liquid chromatography (2D-LC) is being increasingly used for monitoring a variety of CQAs. Chromatography modes such as ion-exchange (IEX), size-exclusion chromatography (SEC), and hydrophilic interaction liquid chromatography (HILIC) are the preferred choices for the first dimension because of their MS-incompatible buffer conditions, whereas chromatography with organic solvent system such as HILIC or reverse-phase liquid chromatography (RPLC) are used in the second dimension to couple with MS. Although these approaches have already been demonstrated to be extremely effective, the use of RPLC or HILIC in the second dimension results in a major limitation in that the variants cannot retain their native form because of the use of organic solvents. This is why mAb variants are conventionally analyzed by conventional analytical IEX or HILIC methods under non-volatile salt-based buffer conditions. However, this approach requires manual fraction collection and desalting steps before analysis with mass spectrometry can be performed. Introduction of a volatile-salt–based mobile phase overcomes the limitation of direct coupling of analytical separation techniques with mass spectrometry. In a recent study, an iMAM method was presented in which an online workflow of a 2D-LC method using an MS-compatible mobile phase coupled with native mass spectrometry was performed to characterize hydrophobic variants in the first dimension and charge variants in the second dimension without needing manual fractionation (Figure 1) (14). As a result of this novel workflow, characterization of 10 variants for one mAb can be achieved, out of which a pair of variants was exclusive to the orthogonal 2D method. Likewise, characterization of a total of 11 variants of a second mAb could be achieved, including five variants that were exclusive to the orthogonal 2D workflow. In comparison, the standalone HILIC method could identify only four variants for both the mAbs, whereas a standalone weak cation-exchange (WCX) method was able to identity seven variants for one mAb and six variants for the other mAb. In addition, common modifications that usually are co-eluted, or at best, are inadequately separated from the main product in standalone HILIC or WCX, could be separated and estimated in the proposed workflow.
FIGURE 1: Schematic representation of a 2D-HIC-WCX MAM workflow which analysis hydrophobic variants in 1D and charge variants analysis in the 2D using native MS.
Case Study II: Monitoring Titer and Size Variants
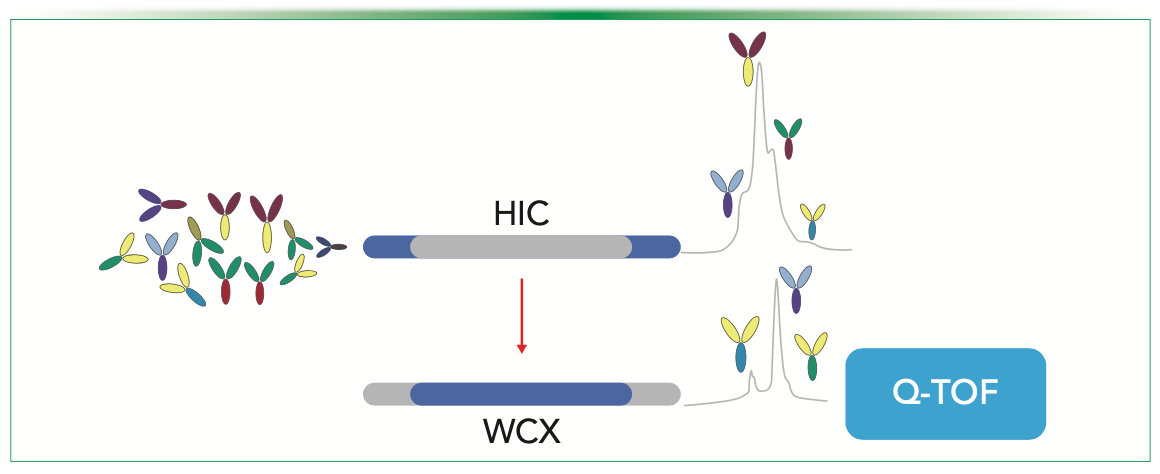
Titer, aggregates, and intact mass are critical attributes to monitor during process development or optimization. Here, we propose an online 2D-LC MAM workflow that combines protein-A affinity in the first dimension and SEC chromatography in the second dimension coupled with MS for the estimation of titer, size heterogeneity, intact mass, and major glycoforms of a mAb (Figure 2). Although estimation of titer and aggregates using a 2D-LC approach have been established (15), the researchers used non-volatile buffer systems with UV detector, lacking the in-depth characterization of size variants that can be achieved by mass spectrometry. In this study, volatile-salt–based chromatography was performed in both dimensions, given the multiple advantages it offers, including reduced ion suppression, faster analysis, fewer desalting stage artifacts, and improved ionization, resulting in more precise analyte identification. In addition, volatile-salt–based chromatography can enhance peak shape and resolution, thereby preventing column fouling and resulting in improved reproducibility of chromatographic separations. In the published study, three major glycovariants were identified for the first mAb, and four major glycovariants for the second mAb, with a separation time of 8 min and using a small sample amount (10–15 μg) without the need for any manual manipulations. Typically, a standalone SEC analysis takes up to 2–3 h for peak collection of ProA followed by buffer exchange to a MS-compatible buffer with the risk of sample loss, degradation, induced modifications, and manual handling, which is likely to negatively impact reproducibility. The proposed method offers applications in quality control (QC) testing, as well as for process analytical technology (PAT) applications in upstream and midstream processing.
FIGURE 2: Schematic representation of 2D-ProA-SEC MAM workflow in which the estimation of titer in 1D and size variants analysis in the 2D using native MS.
Case Study III: Monitoring of Aggregates and Charge Variants
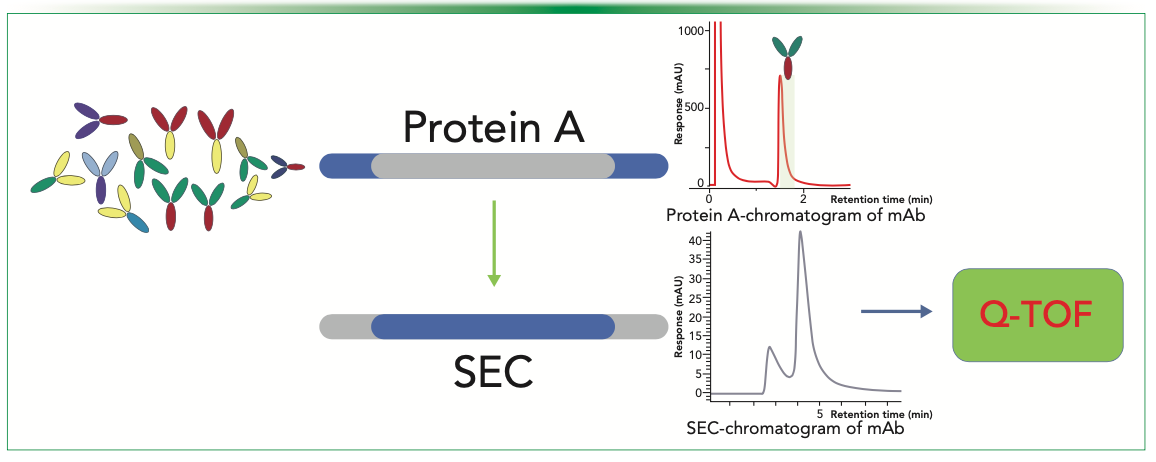
Assessment of CQAs (such as size and charge-related changes) is a critical need for biopharmaceutical manufacturers. Although SEC is widely used for assessment of fragments and aggregates in the product, weak-cation exchange chromatography (WCX) is frequently employed for analyzing charge variants of mAbs. Integration of a 2D-LC method with MAMs offers a more comprehensive and in-depth analysis of complex protein like monoclonal antibodies. Most published MAMs utilize a volatile-salt–based 2D-LC workflow in which SEC chromatography is performed in the first dimension (1D) and WCX chromatography in the second dimension (2D). However, researchers have reported a 2D-LC method for aggregate analysis by a SEC-UV method in 1D and charge variant identification in 2D through a CEX-MS method (16). In the standard 2D-LC workflow, direct analysis through mass spectrometry is limited to the second-dimension samples, as the first dimension is not readily coupled with mass spectrometry. In a recent paper, a novel MAM workflow that enables direct coupling of both chromatography dimensions (1D and 2D) with mass spectrometry has been proposed (Figure 3). This workflow utilizes a volatile-salt based mobile phase in both chromatography steps to achieve efficient ionization and vaporization of the analytes so as to prevent interference with the second-dimension separation and maintain the sensitivity and resolution of the overall analytical method. The proposed approach eliminates the need for manual intervention and facilitates the detection of low-abundance variants. Additionally, this method utilizes 75% less sample and takes less time to analyze (25 min as opposed to 90 min) compared to the separate analyses of size and charge variants using standalone SEC and WCX methods.
FIGURE 3: Schematic representation of 2D-SEC-WCX MAM workflow which analysis size variants in 1D using SEC-MS and charge variants analysis in the 2D using WCX-MS.
Conclusions
Traditional MAM workflows are being increasingly used to enhance biopharmaceutical analysis during production and quality control. By utilizing a bottom-up approach, traditional MAMs enable accurate identification and quantification of most quality attributes with residue specificity. The iMAM workflow, on the other hand, is a suitable alternative that can complement standard MAMs or be used when there is a need for a rapid turnaround time and monitoring of only a limited number of CQAs. The iMAM workflows presented here rely on analyzing the intact mass of mAbs in their native form, resulting in significant improvements in high-throughput sample analysis. These simplified setups and automated data processing could be utilized for various purposes, such as monitoring and comparing process and product quality attributes. Hence, iMAMs could prove to be highly beneficial for high-throughput screening, aiding in process development or for routine monitoring of product quality. Furthermore, iMAMs can be employed in situations where the product is still in culture media or to circumvent extensive sample preparation that may modify certain attributes. Future advances in the workflow may include assessing parameters like linearity and the limit of detection for specific features, as well as implementing system suitability tests that align with the chromatographic approach of interest. Another possible advancement is the use of advanced data analysis techniques such as machine learning and artificial intelligence to analyze the complex data generated by MAMs and 2D-LC. These techniques can help identify patterns and correlations between different product attributes, leading to a deeper understanding of the product’s composition and performance.
References
(1) Fekete, S.; Guillarme, D.; Sandra, P.; Sandra, K. Chromatographic, Electrophoretic, and Mass Spectrometric Methods for the Analytical Characterization of Protein Biopharmaceuticals. Anal. Chem. 2016, 88 (1), 480–507. DOI: 10.1021/acs.analchem.5B04561
(2) Rogers, R. S.; Nightlinger, N. S.; Livingston, B.; Campbell, P.; Bailey, R.; Balland, A. Development of a Quantitative Mass Spectrometry Multi-Attribute Method for Characterization, Quality Control Testing and Disposition of Biologics. MAbs 2015, 7 (5), 881–890. DOI: 10.1080/19420862.2015.1069454
(3) Rogers, R. S.; Abernathy, M.; Richardson, D. D.; Rouse, J. C.; Sperry, J. B.; Swann, P.; Wypych, J.; Yu, C.; Zang, L.; Deshpande, R. A View on the Importance of “Multi-Attribute Method” for Measuring Purity of Biopharmaceuticals and Improving Overall Control Strategy. AAPS J. 2017, 20 (1). DOI: 10.1208/S12248-017-0168-3
(4) Auclair, J. R.; Rathore, A. S. The Multi-Attribute Method (Mam) for the Characterization of Biopharmaceuticals. LCGC North Am. 2021, 39 (1), 28–32. DOI: 10.56530/lcgc.na.GI5577L2
(5) Martelet, A.; Garrigue, V.; Zhang, Z.; Genet, B.; Guttman, A. Multi-Attribute Method Based Characterization of Antibody Drug Conjugates (ADC) at the Intact and Subunit Levels. J. Pharm. Biomed. Anal. 2021, 201, 114094. DOI: 10.1016/j.jpba.2021.114094
(6) Carillo, S.; Criscuolo, A.; Füssl, F.; Cook, K.; Bones, J. Intact Multi-Attribute Method (IMAM): A Flexible Tool for the Analysis of Monoclonal Antibodies. Eur. J. Pharm. Biopharm. 2022, 177, 241–248. DOI: 10.1016/j.ejpb.2022.07.005
(7) Sitasuwan, P.; Powers, T. W.; Medwid, T.; Huang, Y.; Bare, B.; Lee, L. A. Enhancing the Multi-Attribute Method through an Automated and High-Throughput Sample Preparation. MAbs 2021, 13 (1). https://doi.org/10.1080/19420862.2021.1978131
(8) Evans, A. R.; Hebert, A. S.; Mulholland, J.; Lewis, M. J.; Hu, P. ID-MAM: A Validated Identity and Multi-Attribute Monitoring Method for Commercial Release and Stability Testing of a Bispecific Antibody. Anal. Chem. 2021, 93 (26), 9166–9173. DOI: 10.1021/acsS.analchem.1C01029
(9) Rogers, R. S.; Abernathy, M.; Richardson, D. D.; Rouse, J. C.; Sperry, J. B.; Swann, P.; Wypych, J.; Yu, C.; Zang, L.; Deshpande, R. A View on the Importance of “Multi-Attribute Method” for Measuring Purity of Biopharmaceuticals and Improving Overall Control Strategy. AAPS J. 2018, 20 (1), 1–8. DOI: 10.1208/S12248-017-0168-3
(10)“Quality Considerations for the Multi-Attribute Method (MAM) for Therapeutic Proteins - 10/13/2022,”| FDA. https://www.fda.gov/science-research/fda-grand-rounds/quality-considerations-multi-attribute-method-mam-therapeutic-proteins-10132022 (accessed 2023-03-07).
(11) Millán-Martín, S.; Jakes, C.; Carillo, S.; Buchanan, T.; Guender, M.; Kristensen, D. B.; Sloth, T. M.; Ørgaard, M.; Cook, K.; Bones, J. Inter-Laboratory Study of an Optimised Peptide Mapping Workflow Using Automated Trypsin Digestion for Monitoring Monoclonal Antibody Product Quality Attributes. Anal. Bioanal. Chem. 2020, 412 (25), 6833–6848. DOI: 10.1007/S00216-020-02809-Z/
(12) Liu, P.; Zhu, X.; Wu, W.; Ludwig, R.; Song, H.; Li, R.; Zhou, J.; Tao, L.; Leone, A. M. Subunit Mass Analysis for Monitoring Multiple Attributes of Monoclonal Antibodies. Rapid Commun. Mass Spectrom. 2019, 33 (1), 31–40. https://doi.org/10.1002/RCM.8301
(13) Lanter, C.; Lev, M.; Cao, L.; Loladze, V. Rapid Intact Mass Based Multi-Attribute Method in Support of MAb Upstream Process Development. J. Biotechnol. 2020, 314–315, 63–70. https://doi.org/10.1016/J.Jbiotec.2020.04.001
(14) Sarin, D.; Kumar, S.; Rathore, A. S. Multiattribute Monitoring of Charge-Based Heterogeneity of Recombinant Monoclonal Antibodies Using 2D HILIC-WCX-MS. Anal. Chem. 2022, 94 (43), 15018–15026. https://doi.org/10.1021/ACS.analchem.2C02931
(15) Dunn, Z. D.; Desai, J.; Leme, G. M.; Stoll, D. R.; Richardson, D. D. Rapid Two-Dimensional Protein-A Size Exclusion Chromatography of Monoclonal Antibodies for Titer and Aggregation Measurements from Harvested Cell Culture Fluid Samples. MAbs 2020, 12 (1). https://doi.org/10.1080/19420862.2019.1702263
(16) Lambiase, G.; Inman, S. E.; Muroni, M.; Lindo, V.; Dickman, M. J.; James, D. C. High-Throughput Multiplex Analysis of MAb Aggregates and Charge Variants by Automated Two-Dimensional Size Exclusion-Cation Exchange Chromatography Coupled to Mass Spectrometry. J. Chromatogr. A 2022, 1670. https://doi.org/10.1016/J.Chroma.2022.462944
About the Co-Author
Sunil Kumar is a postdoctoral fellow working with Prof. Anurag S. Rathore in the Department of Chemical Engineering at the Indian Institute of Technology in Delhi, India.

About the Author
Jared Auclair is an Associate Dean of Professional Programs and Graduate Affairs at the College of Science at Northeastern University, in Boston, Massachusetts. He is also the Director of Biotechnology and Informatics, as well as the Director of the Biopharmaceutical Analysis Training Laboratory.

About the Column Editor
Anurag S. Rathore is a professor in the Department of Chemical Engineering at the Indian Institute of Technology in Delhi, India.


Study Explores Thin-Film Extraction of Biogenic Amines via HPLC-MS/MS
March 27th 2025Scientists from Tabriz University and the University of Tabriz explored cellulose acetate-UiO-66-COOH as an affordable coating sorbent for thin film extraction of biogenic amines from cheese and alcohol-free beverages using HPLC-MS/MS.
Quantifying Microplastics in Meconium Samples Using Pyrolysis–GC-MS
March 26th 2025Using pyrolysis-gas chromatography and mass spectrometry, scientists from Fudan University and the Putuo District Center for Disease Control and Prevention detected and quantified microplastics in newborn stool samples.












