Impurity Determinations for Biotechnology-Derived Biopharmaceuticals and Related Products
LCGC North America
This installment of "Validation Viewpoint" column addresses, in the hopes of clarifying what the biopharmaceutical industry is required to do today to identify and quantify impurities in their biotech, proteinaceous products.
In pharmaceutical production of small molecule synthetic drugs, it is an FDA requirement that the manufacturer demonstrate the presence or absence of impurities at or above the 0.1% level (1–2). That level refers to the high performance liquid chromatography (HPLC)–UV peak areas of the suspected impurities in comparison to the total peak areas for all drug-related substances present in a final drug substance or drug formulation. For each impurity present, the structure and toxicity must be firmly demonstrated to the FDA's satisfaction. If the impurity demonstrates harmful side effects, such as radical toxicity, then it must be controlled in the final drug formulation (1–2). The isolation, purification, and characterization of synthetic drug impurities are routinely performed.

Michael E. Swartz
However, the same cannot always be said of impurities present in biopharmaceutical- or biotechnology-derived commercial products. It is this issue that this "Validation Viewpoint" column addresses, in the hopes of clarifying what the biopharmaceutical industry is required to do today to identify and quantify impurities in their biotech, proteinaceous products. Perhaps the most commonly encountered impurities in biotech products are host cell-derived, also termed host cell impurities (HCIs). Of these, the most common are host cell proteins (HCPs) or host cell DNA (HCDNA). There are several currently accepted analytical methods to detect and characterize both HCPs and HCDNA. The nature of these impurity species is clearly dependent upon the particular host cell expression system, whether yeast, bacterial, or mammalian.

Ira S. Krull
In doing impurity determinations for biotech products, it is often impossible to characterize or quantitate the impurities fully. This inability to characterize biotechnology-derived product impurities presents a significant challenge requiring a much different approach.
Why are Biotechnology Impurities Difficult to Characterize and Quantitate?
In general, host cell-derived biotechnology impurities are not of the very same amino acid sequence as the biopharmaceutical substance itself. Sometimes variants or isoforms of the bulk biopharmaceutical, which can contain different glycosylation patterns or contents, or might have an amino acid transformation (for example, glutamic acid to pyroglutamate), an amino acid deletion (for example, loss of a C-terminal lysine group), or other commonly and easily identified posttranslational modifications (PTMs) are encountered (4–12). Such variants most commonly have different isoelectric points (pI), molecular weights (MWs), amino acid content, or peptide map, but they almost always have a basic amino acid backbone with relatively minor PTMs. Even aggregates are considered variants of the monomer itself, often having the same activity towards a binding partner but often different immunogenicity properties. However, PTMs are not the same things as impurities of the drug substances. Impurities can be HCP, HCDNA pieces, or other nucleic acids coming from the host cell itself, or newer proteins coming by recombination of smaller peptides of the original proteome after proteolysis in the cell (protein reorganizations). Impurities can be clipped fragments of the intact, recombinant protein of varying residue lengths. Other impurities can arise as a result of processing steps in the purification train for the drug substance itself, such as protein A leaked from an affinity column used to purify the target drug substance. There are an almost infinite number of possible impurities, most of which might be known but not all.
So, perhaps a first question becomes: Which peaks in the HPLC chromatogram of the intact protein are impurities and which are PTMs or variants and are desired or allowed in the final drug formulation? Second: How does one differentiate between these two basic groups of HPLC peaks — protein variants (desired) and impurities (undesired and to be identified or removed from the final formulation)? True variants are active against a certain target (biologically active species), and usually that target is involved in a disease process, be it a small molecule, peptide, protein, or antibody. Quite often, the basic biopharmaceutical is a mixture of variants, but all are (usually) active against the same (usually) target. In the case of antibody drugs, they all have basically the same epitopic (antigen binding domain) binding or complementary determining region (CDR) and recognize the same antigen or antibody (9,10). One could define a variant as an isoform of the basic protein that binds to its target or receptor, and an impurity as something that does not.
The difference between variants and impurities often can be demonstrated readily by the use of immunoassays, for example, enzyme-linked immunosorbent assays (ELISAs) or others, as well as by competitive binding assays using chromatography, electrophoresis, surface plasmon resonance (SPR), and related techniques (10,11,13–17). It also is possible that there are other definitions of an impurity in a biopharm product, but if a peak in a chromatogram of all species present does not respond to its antigen or target, then it cannot be considered to be an integral part of the product or drug formulation (excluding excipients, sugars, polyols, and polyethylene glycols). Rather, impurity proteins are species somehow related to the true drug substance and product, but which are not active against the desired target or receptors. HCDNA species often contaminate the protein drug by leaching from the cell when lysed, and though not at all proteinaceous, they are still related, indirectly, to the drug species.
Biotechnology-derived impurities can be very difficult to characterize and quantify, because they often are present at very low levels, and because they can represent very complicated species or mixtures of species. It is also very difficult to obtain an authentic reference standard of the impurity peaks (other than for something such as protein A). Thus, FDA requirements today usually only require identification as is possible and relative quantitation using relative peak areas in comparison to all other HPLC peaks (for example, present in the final drug substance). However, to fully characterize a trace amount of an impurity protein, perhaps host cell derived, without having reference materials or even a database (protein searching database, such as Mascot, Prowl, or SwissProt) becomes a time consuming, lengthy, and often very expensive process. It is orders of magnitude more difficult than isolating, characterizing, and quantifying a synthetic drug impurity in a conventional pharmaceutical substance. Of course, the HPLC or high performance capillary electrophoresis (HPCE) chromatograms or electropherograms of HCIs, as well as their mass spectra, usually are different than those for the target drug substance itself. Running cell expression blanks, without generating the drug substance, often makes it easier to discern what HCIs are present and then how to separate these from the drug substance itself (18–20).
Methods for the isolation of HCIs rely on the standard, analytical, or preparative methods for purification of the drug substances, mainly HPLC at various levels. Indeed, purification of the drug substance often provides enough HCIs for isolation and characterization. The characterization of HCPs is done using exactly the same analytical methods used at the very same time to characterize the parent drug substances (4–9, 20). Figure 1 illustrates some typical HPLC (ion-exchange) chromatograms derived using various commercial columns to resolve HCPs from the drug substance itself (20). Using four different HPLC columns, it is clear that some columns, such as DEAE-FF (Figure 1d), are better able to resolve HCP(s) from the protein of interest and other unknown, host cell-derived species. A totally different order of elution is illustrated in Figure 1c for a Q-FF column.
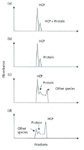
Figure 1: Series of HPLC chromatograms for various columns and their ability to resolve HCPs from the drug substance itself (20). (Reprinted with permission of the copyright holder, Elsevier Science Publishers and the Journal of Pharmaceutical and Biomedical Analysis.)
Characterization of HCDNAs, however, requires the development and optimization of standard DNA procedures, again mainly HPLC-based, such as ion-exchange chromatography, paired-ion reversed-phase HPLC, flat-bed electrophoresis, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and SDS–capillary gel electrophoresis (CGE), (21–25).
Capillary isoelectric focusing (cIEF) has been used for the separation and characterization of protein process (not HCPs) impurities in a typical recombinant protein product (18,19). Figure 2 illustrates the application of cIEF for these "impurities" in this protein product. In this particular example, three product variants (protein process impurities) are resolved successfully (Figure 2a), migrating after the drug substance itself, the fastest migrating species. Figure 2b illustrates the use of (external) protein markers to determine the approximate pIs of the species in Figure 2a. However, a calibration plot for pI measurements must be done using the reference proteins present in the very same injection (and sample) of those species whose pI is being determined. Otherwise, incorrect pI values will almost always result.
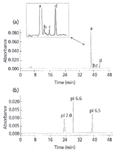
Figure 2: cIEF electropherogram of a typical analysis of a recombinant protein product containing numerous variants or isoforms (19). (Reprinted with permission of the copyright holder, Elsevier Science Publishers and the Journal of Chromatography, A.)
There is also, perhaps, a question of semantics in defining host cell impurities versus drug substance impurities or variants. Though today, ICH can define protein impurities as being variants, really PTMs, of the drug substance itself, this approach is misleading (26). Product-related impurities for protein biotechnology products are described in the ICH guidelines as molecular variants that arise from processing or during storage. These are, of course, very different from HCIs, which is the topic of discussion here. A case can be made that the ICH guidelines are incorrect in equating protein–drug variants as being product related impurities. Variants of the parent drug are variants or isoforms, not true impurities, and are a fundamental part of the total drug substances that need not be removed, just characterized.
Common Methods for Purification and Isolation of HCIs
HCIs also have been termed host cell contaminants at times. In addition to HCDNA and HCPs, other possible impurities are often endotoxins, as well as biomolecules normally found in the expression system needed for cell survival, often smaller, lower molecular weight molecules. In addition to the most commonly used isolation methods described previously, HPLC (size-exclusion, cation-exchange, and hydrophobic interaction chromatography) and HPCE, centrifugation and precipitation also are used often. In addition to HPCE, it is common to use 1D and 2D gels, Western blotting, antigen–antibody recognition, and other standard analytical techniques. In the final analysis, all of these techniques only resolve or isolate the HCPs from the drug substance and nucleic acids, as in Figure 1. They do not identify the HCPs, and for that purpose, mass spectrometry (MS) and MS-MS have become standard analytical techniques in common use today in all biotechnology companies.
As mentioned earlier, isolation of HCDNA species commonly is done also using ion-exchange HPLC and SDS-PAGE or SDS-CGE methods to detect the presence of HCDNA, immunoassays (complementary DNA binding) are often used, as well as the DNA threshold test, perhaps together with polymerase chain reaction (PCR) to amplify any HCDNA present in the purified drug substance or drug products (27). The absolute identify of the HCDNA species, once separated, can be performed using complementary DNA binding assays, MS and MS-MS, or DNA sequencing using SDS-CGE or other methods. Figure 3 illustrates, in diagrammatic fashion, the basic DNA threshold test, using a single-stranded (ss) DNA-binding protein that collects the suspect ssDNA to a membrane, and an antibody–enzyme that recognizes the ssDNA and is then able to generate substrate product turnover (urease causing urea to form ammonia and carbon dioxide, measured electrochemically or by conductivity) (20,27).
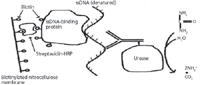
Figure 3: Assay mechanism to quantitate host cell derived DNA (20). (Reprinted with permission of the copyright holder, Elsevier Science Publishers and the Journal of Pharmaceutical and Biomedical Analysis.)
Absolute Quantitation of Protein Impurities by Currently Available, Commercial Software
One of the key validation requirements for both small and large molecule pharmaceuticals has to do with accuracy of quantitation (3,28–30). Accuracy here refers to demonstrating that the amount or level of a protein being often quantitated in an absolute (not relative) sense agrees with the actual or true levels to be found in those samples. Accuracy requires performing either relative or absolute quantitations of the drug substance or HCPs or other HCIs. With synthetic drugs, determining accuracy is usually an easy matter, as authentic reference materials for the drug substance and its impurities are simple to synthesize or purchase from commercial vendors. However, in the case of HCPs, less so than for drug substance variants, obtaining reference materials becomes more and more difficult. Most expression system HCPs already are known, and often, these can be purchased in fairly high purities. But this is usually not the case for drug substance variants, especially for complex ones such as glycosylated or phosphorylated isoforms. Adding to the challenge, it is not always a simple matter to synthesize or purchase authentic standards for such variants. Thus, relative quantitation using percent peak areas has become de rigeur in the biopharmaceutical industry, which does not agree with absolute levels present necessarily (concentrations, for example, milligrams per milliliter). However, there are some techniques on the market today that will permit absolute quantitations for even protein variants of incompletely proven structures (for example, glycoproteins).
The accuracy of an analytical procedure "expresses the closeness of agreement between the value which is accepted either as a conventional true value or an accepted reference value and the value found" (28–30). It is commonly estimated by measuring the recovery of known amounts of the test substance, after spiking into blank matrices. For impurity methods, recovery is measured typically at three concentrations that span the expected impurity content of a sample. In the case of protein impurities, however, it is often impractical to spike the impurity into a blank matrix because this usually will result in significant sample loss at low impurity concentrations due to adsorption on contact surfaces like the capillary, sample vial. It is possible to blend each of the two impurity species separately with the drug substance. Theoretical percent impurity then can be calculated from protein concentration of the parent protein and impurity species, and the mixing ratio of the two species. This calculation assumes that the UV absorption properties (for example, extinction coefficients) of all proteins involved are about the same.
In a recent study, Rathore and colleagues (20) looked at six impurity levels that were generated over the range of interest, and recovery of the impurity was determined by comparing the measured area percent for each impurity peak versus the theoretical percent impurity. In this study, drug substance variants were studied for recovery of the deamidated and aggregated impurity species, respectively. Three capillary isoelectric focusing (cIEF) analyses were performed for each impurity level. The averaged area percent values for spiked samples were corrected by subtracting the peak area percent values for the unspiked drug substance. The corrected area percent value was compared to the theoretical percent impurity (based upon the blending ratio) for each spike level and the ratio of these two quantities was reported as recovery. Data were used to compute repeatability (RSD values) and accuracy (measured area percent versus theoretical percent impurity). For both impurity species, recovery was lowest at the higher impurity levels. For the deamidated impurity, recovery was >90% for the 0.5–2% impurity levels, and >85% for impurity levels of 3–12%. For the aggregated impurity, the recovery improved to 97 % as the impurity level increased from 0.5 % to 3%, and then abruptly declined to <80% for impurity level >6%. With the exception of the 73% recovery for the highest aggregated impurity level of 8%, the recovery values were >80%. The study is really a variation of the standard additions approach to absolute quantitation, so long used in inorganic analysis (31).
The "standard addition approach" discussed earlier is a better-than-typical approach to determining absolute quantitation and accuracy of protein variants, though not yet for HCPs. In the future, another as yet untried, absolute quantitation approach can involve high-resolution MS together with certain newer software (Expression software,Waters Corporation, Milford, Massachusetts) now available from at least two commercial vendors (32,33). Using such instrumentation and software, it is possible with suitable internal standards unrelated to the drug substance variants or HCPs, to determine absolute concentrations present for virtually any protein samples without using recovery or standard additions-type approaches. These newer methods of protein quantitation require an initial enzymatic digestion of all proteins of interest into the usual peptide maps, and then HPLC-electrospray ionization (ESI)–MS-MS to identify the peptides (and from them their parent proteins) and to perform absolute quantitations on the basis of comparing the largest peptides present for each protein with the internal standard peptide maps. This approach, though somewhat new, has been shown to provide accurate and precise protein levels, even in very complex proteomics-type samples. There is every reason to suspect that these very same analytical methods immediately would be applicable to virtually any commercial, biotechnology product of a proteinaceous nature. Hopefully, this technology will all come about in the near future, and biotechnology analysts will then be able to generate absolute quantitations of the drug substances, its variants and all HCPs, even without performing immunoassays for those HCPs.
Conclusions
For proteins produced in the biotechnology industry, complementary separation techniques are necessary both to purify the target protein and to give an accurate picture of the quality of the final product. The complexity of the product eliminates the use of simple one-dimensional separation strategies. Host-cell contaminants include the full range of molecules present in living cells. During purification and analysis, special concern is given to the separation and detection of host-cell DNA and host-cell proteins. To ensure high quality, yield often is sacrificed for purity in the purification of the therapeutic protein: purity is achieved only by multiple separations based upon differing selectivities. Three analytical methods often are necessary to provide a complete assessment of quality with regard to HCIs. The content of the protein product is determined by HPLC using a variety of modes (for example, see Figure 1) — size-exclusion, ion-exchange, and reversed-phase modes all would be appropriate, but reversed phase is the most common. Host-cell DNA is quantified by DNA threshold methods, and a process-specific immunoassay can quantify extremely low levels (1 ng/mL or 10 ppm) of HCPs. It is this last HCP assay that contributes to the "process defines product" aspect of quality control for biologics. Immunoassays also contribute heavily to the time and cost of quality assessments in biopharmaceuticals. If one could eliminate such immunoassays, as suggested earlier, a tremendous amount of time, effort, money, and materials potentially could be saved.
Michael E. Swartz
"Validation Viewpoint" Co-Editor Michael E. Swartz is Research Director at Synomics Pharmaceutical Services, Wareham, Massachusetts, and a member of LCGC's editorial advisory board.
Ira S. Krull
"Validation Viewpoint" Co-Editor Ira S. Krull is an Associate Professor of chemistry at Northeastern University, Boston, Massachusetts, and a member of LCGC's editorial advisory board..
The columnists regret that time constraints prevent them from responding to individual reader queries. However, readers are welcome to submit specific questions and problems, which the columnists may address in future columns. Direct correspondence about this column to "Validation Viewpoint," LCGC, Woodbridge Corporate Plaza, 485 Route 1 South, Building F, First Floor, Iselin, NJ 08830, e-mail lcgcedit@lcgcmag.com
References
(1) International Conference on Harmonization QATAR(R): Impurities in New Drug Substances, October 2006.
(2) International Conference on Harmonization AB(R): Impurities in New Drug Products, June 2006.
(3) I.S. Krull and M.E. Swartz, LCGC 16(3), 258 (1998).
(4) G. Marko-Vargy and P. Oroszlan, Emerging Technologies in Protein and Genomic Material Analysis (Elsevier Science Publishers, Amsterdam, The Netherlands, 2003).
(5) R. Rodriguez-Diaz, Analytical Techniques for Biopharmaceutical Development (Marcel Dekker, New York, 2005).
(6) C.T. Walsh. Posttranslational Modification of Proteins, Expanding Nature's Inventory (Roberts and Company, Publishers, Englewood, Colorado, 2006).
(7) A.R. Mire-Sluis, Ed., State-of-the-Art Analytical Methods for the Characterization of Biological Products and Assessment of Comparability, Developments in Biologicals, Volume 122 (Karger Basel, Switzerland, 2005).
(8) S.J. Higgins and B.D. Hames, Eds., Post-Translational Processing, A Practical Approach (Oxford University Press, New York, 1999).
(9) L.C. Santora, I.S. Krull, and K. Grant, Anal. Biochem. 275, 98 (1999).
(10) L.C. Santora, P. Sakorafas, Z. Kaymakcalan, I.S. Krull, and K. Grant, Anal. Biochem. 299(2), 119 (2001).
(11) I.S. Krull, S. Kazmi, H. Zhong, and L.C. Santora, Separation of Glycoproteins by cIEF, in Methods in Molecular Biology, Volume 213: Capillary Electrophoresis Separation of Carbohydrates, P. Thibault and S. Honda, Eds. (The Humana Press, Inc., Totowa, New Jersey, 2002), pp. 197.
(12) L.C. Santora, I.S. Krull, and K. Grant, Spectroscopy 17(5), 50 (2002).
(13) I.S. Krull, J. Dai, C. Gendreau, and G. Li, J. Pharm. Biomed. Anal. 16, 377 (1997).
(14) J. Dai, G. Li, and I.S. Krull, J. Pharm. Biomed. Anal. 17, 1143 (1998).
(15) G. Li, X. Zhou, Y. Wang, A. El-Shafey, N.H.L. Chiu, and I.S. Krull, J. Chromatography 1053(1/2), 253 (2004).
(16) H. Zou, Y. Zhang, P. Lu, and I.S. Krull, Biomed. Chromatogr. 10, 78 (1996).
(17) A. El Shafey, G. Jones, H. Zhong, and I.S. Krull, Electrophoresis 23(6), 945 (2002).
(18) A.S. Rathore, R.R. Kurumbail, and A.M. Lasdun, LCGC 20(11), 1042–1050 (2002).
(19) A.M. Lasdun, R.R.Kurumbail, N.K. Leimgruber, and A.S. Rathore, J. Chrom. A, 917, 147–158 (2001).
(20) A.S. Rathore, S.E. Sobacke, T.J. Kocot, D.R. Morgan, R.L. Dufield, and N.M. Mozier, J. Pharm. Biomed. Anal. 32, 1199–1211 (2003).
(21) E.D. Katz, Ed., High Performance Liquid Chromatography: Principles and Methods in Biotechnology (John Wiley & Sons, Inc., New York, 1996).
(22) P. Camilleri, Ed., Capillary Electrophoresis Theory and Practice (CRC Press, Boca Raton, Florida, 1993).
(23) R. Weinberger, Practical Capillary Electrophoresis, Second Edition (Academic Press, New York, 2000).
(24) B.L. Karger and W.S. Hancock, Eds., High Resolution Separation and Analysis of Biological Macromolecules, Part A, Fundamentals (Academic Press, New York, 1996).
(25) P.G. Righetti, Ed., Capillary Electrophoresis in Analytical Biotechnology (CRC Press, Boca Raton, Florida, 1996).
(26) ICH-Topic Q6B: Specifications: Test procedures and acceptance criteria for biotechnological/biological products (International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use, Geneva Switzerland, 1998).
(27) J. Briggs and P.R. Panfili, Anal. Chem. 63, 850–859 (1991).
(28) Draft Guidance on Specifications: Test Procedures and Acceptance Criteria for Biotechnological/Biological Products (FDA, CBER, ICH), 24 July 1998.
(29) Position Statement on the Use of Tumorigenic Cells of Human Origin for the Production of Biological and Biotechnoogical Medicinal Products. Committee for Proprietary Medicinal Products (CPMP), 1 March 2001, CPMP/BWP/1143.
(30) I.S. Krull and M.E. Swartz, LCGC 23(1), 46 (2005).
(31) G.D. Christian. Analytical Chemistry, Sixth Edition (John Wiley & Sons, Inc., New York, 2004).
(32) J.C. Silva, R. Denny, C. Dorschel, M.V. Gorenstein, G.-Z. Li, K. Richardson, D. Wall, and S.J. Geramanos, Molecular & Cell. Proteomics, 5, 589–607 (2006).
(33) H. Vissers, M. Kipping, T. Reimer, A. Kasten, C. Koy, J. Langridge, and M. Glocker, Determination of the Quantitative Protein Signatures for Ductal Carcinoma (Breast Cancer) by LC–MS Proteome analysis, Part I, Waters Corporation, Application Note, 2007.













Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)