Efficient Quantitation of Biotherapeutics in Biological Matrices Through the Application of LC–MS‑Related Techniques
The Column
Biotherapeutics must endure in-depth testing to validate their efficacy and safety before their release to the medical community. Characterization and quantitation of these large molecule medicines is traditionally performed with ligand binding assays or radiolabeling procedures. Issues with selectivity, accuracy, and unavailability of applicable assays for the characterization and quantitation of certain biotherapeutics means that liquid chromatography–mass spectrometry (LC–MS) is becoming an increasingly selected method for biotherapeutics testing. Typically used for small molecules, LC–MS can be adapted for larger molecule analysis with additional high throughput and multiplexing capabilities. New method development has turned LC–MS into a highly sensitive option for biotherapeutics validation.
Photo Credit: kentoh/Shutterstock.com

Debadeep Bhattacharyya1, Bo An2, and Jun Qu2, 1Thermo Fisher Scientific, Cambridge, Massachusetts, USA, 2Depart. Pharm. Sci., SUNY-Buffalo, Buffalo, New York, USA
Biotherapeutics must endure in-depth testing to validate their efficacy and safety before their release to the medical community. Characterization and quantitation of these large molecule medicines is traditionally performed with ligand binding assays or radiolabeling procedures. Issues with selectivity, accuracy, and unavailability of applicable assays for the characterization and quantitation of certain biotherapeutics means that liquid chromatography–mass spectrometry (LC–MS) is becoming an increasingly selected method for biotherapeutics testing. Typically used for small molecules, LC–MS can be adapted for larger molecule analysis with additional high throughput and multiplexing capabilities. New method development has turned LC–MS into a highly sensitive option for biotherapeutics validation.
Biotherapeutics are a valuable aspect of medicine in the treatment and prevention of disease. These larger, more complex molecules are different from chemically synthesized small molecule drugs, in that they are proteins or peptides derived from living systems including mammalian cells, viruses, or bacteria. As biotherapeutics are made for a diverse range of specific targets as opposed to nonspecific dispersion, they are integral to delivery of cutting-edge treatments. Their structural similarity to endogenous molecules allows biotherapeutics, such as monoclonal antibodies (mAbs) and several protein-based therapeutics, to exhibit high efficacy in many disease areas.
Standard Techniques for Development of Biotherapeutics
The multimodal biotherapeutics production process requires precision, conformity to good manufacturing processes, and defined specifications to maintain the safety and efficacy of the product. As good characterization of products is required before release, numerous in-process tests are necessary for quality assurance. In addition, robust and reliable bioanalytical methods help to generate pharmacokinetic (PK), pharmacodynamic (PD), and immunogenicity data that ensures product safety and efficacy throughout the development process.
Ligand binding assays (LBA) and radiolabeling are common techniques used to analyze biotherapeutic effects and distribution in the human body. The use of ligand binding assays, such as enzyme-linked immunosorbent assay (ELISA) and its variations including fluorescence immunoassay (FIA), chemiluminescence immunoassay (CLIA), time-resolved fluorescence assay (TRF), and electrochemiluminescence assays (ECL), allows users to benefit from high sensitivity and high throughput, as well as excellent reproducibility in plasma-based testing. However, validated methods exist for a limited number of mAbs and new method development is a costly and time-consuming process; moreover, the use of LBAs can be matrix- and assay-selective.
Radiolabeling assays, where biotherapeutic proteins are tagged with radioactive lowâmolecular-weight reagents, are quick and easy to develop and are not matrix specific, but they are limited to animal studies because of radioactive exposure concerns. Selectivity and accuracy of biodistribution assays can also be problematic with radiolabeling being an issue, as well as pharmacokinetic changes caused by the physicochemical features of the labelling reagent.
Liquid chromatography–mass spectrometry (LC–MS) has been the premier tool across the areas of pharmaceutical research, development, and production including metabolism, toxicology, proteomics, and formulation. LC–MS technology offers a promising solution for targeted quantification. The technique allows robust, sensitive, fast, and easy method development in a host of matrices and also provides the high throughput and multiplex capacity to enable productivity goals. Even though LC–MS is primarily used for small molecule evaluation, new method development can adapt the technique to characterize and quantify biotherapeutics. Proteinâbased biotherapeutics are analyzed using signature peptides produced through a controlled digestion process, which can be easily identified and quantified at very low concentrations with LC–MS. Such an analytical process allows unmatched sensitivity and accuracy in the detection and quantitation of the signature peptides that represent the larger biotherapeutic.
Solutions for Sample Preparation Challenges
It is important that initial sample preparation, primarily sample clean-up and digestion, is optimized to monitor the target analyte correctly. For example, drug extraction from tissue samples presents a difficult challenge. Blood must be removed from the tissue to eliminate positive bias from any drug in circulation. Conversely, excessive perfusion can cause a loss of tissue-bound proteins. With no current optimal strategy for blood removal, a differential injection method was developed to efficiently remove plasma and reduce bias. By injecting animals with a high concentration of nonlabelled antibody that infuses into tissues followed hours later by injection of heavy isotope-labelled antibody that remains in the blood as a marker for blood contamination, effective blood removal is possible as is more accurate determination of drug concentration in tissue. The new method was used to direct perfusion efforts, which results in blood removal efficiency of 98–99% and reduces positive bias to less than 5%.
Enzymatic methods of sample clean-up post-digestion can also be challenging, resulting in dirty samples with in-solution digestion or inefficiencies with in-gel digestion. To improve this process, a twoâstep method, surfactant-aided precipitation/onâpellet-digestion (SOD), can be employed using a high concentration of detergents to extract the sample, succeeded by organic precipitation to remove matrix components and the remaining detergents. This method not only provides fast and clean digestion, but also yields high protein recovery and displays good efficiency, sensitivity, and reproducibility (1).
Improving Sensitivity with Nano-LC–MS
When quantifying biotherapeutics, and much like small molecules, sensitivity tends to be a common expectation. High-molecular-weight proteins are harder to analyze and measure, enhanced by difficulty in detecting lower levels in tissue than in plasma. However, it is extremely important to maintain high levels of sensitivity for quantification by decreasing chemical noise from contaminants and other components to detect these low drug concentrations and provide valid safety assessments. High sensitivity can be achieved by enriching the target peptide and moving to an inlet with low-flow rate by modifying LC–MS to nano-LC–MS.
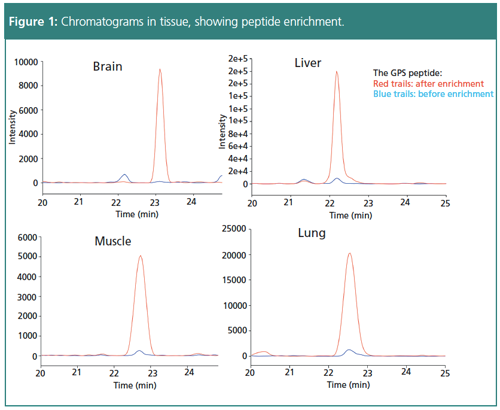
Dual-mechanism cartridge enrichment saves time spent on traditional antibodyâbased peptide enrichment. A double wash initially removes noncharged matrix peptides and then removes polar matrix peptides by changing to a high pH buffer. After selective elution, enrichment removes more than 95% of the matrix peptide while retaining more than 90% of the target peptide. Cartridgeâbased peptide enrichment before LC–MS can result in clean peak signals with quantification limits of 5–10 ng/mL, decreasing chemical noise significantly.
To further improve sensitivity, a novel LC–MS technique utilizing low flow has been developed. Based on the rationale that electrospray ionization (ESI)-MS is concentration-dependent, lower flow would create a higher peak concentration. Essentially, a smaller column could result in a larger signal response. Theoretically, if using a 4.6-mm column results in a 1× signal response and a 0.3-mm column results in a 200× signal response, then a 0.075-mm column would result in a 1600× response. Applying this reasoning, development of nano-LC–MS achieves low limits of detection (LOD) of 20–100 attomole per injection, allowing analysis of minimal drug exposure effects across different peptide signatures.
Optimization of Method Development
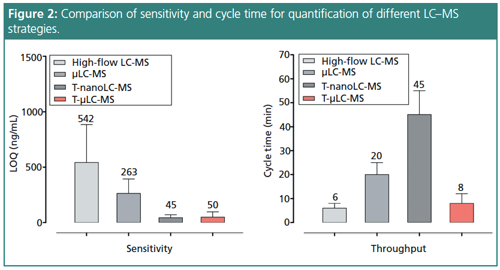
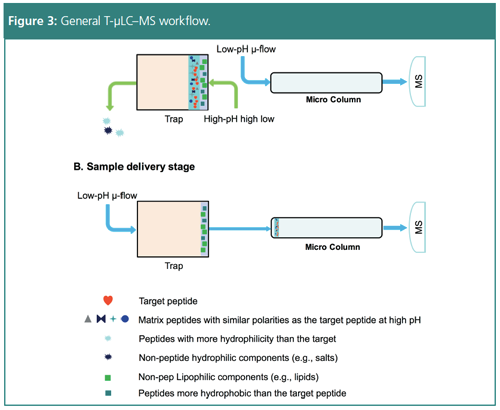
Designing and implementing novel techniques for significant method improvement aids in the performance of quantification efforts. Nevertheless, there are improvements that however useful need further optimization depending on the application. Nano-LC–MS delivers excellent sensitivity but is not robust or high-throughput enough for largeâscale studies common in pharmaceutical development (2,3). In addition, it has a low loading capacity for complex samples and long analytical cycles, all of which reduce productivity (3). In order to scale nano-LC–MS, the method can be adapted to trapping micro-LC–MS (T-µLC–MS). High pH trapping decreases chemical noise and is helpful for large-scale protein analysis, increasing sensitivity across multiple samples.
The selective analysis workflow begins with high flow chromatography at a high pH to trap the sample, forcing out any hydrophilic peptides and matrix components. Switching to a low pH micro-flow isolates the targeted peptides, and a successive high flow removes hydrophobic peptides after switching the trap offline. This process focuses the analysis on a small fraction of sample to improve speed and robustness. Subsequent high-resolution mass spectrometry improves the signalâtoânoise ratio (s/n) by five times compared to standard isolation. T-µLC–MS allows sensitive analysis of thousands of samples with large capacity peptide loading, making it as sensitive as nano-LC–MS and as high throughput and robust as standard LC–MS.
Further optimization of the method using on-the-fly orthogonal array optimization for selected reaction monitoring (SRM) in tandem MS provides multiplexed evaluation of peptide stability and sensitivity. Peptide stability can often be overlooked but is an important aspect of optimization procedures, potentially causing failure of protein validation. This optimization method enables selection of the best signature peptides for a specific protein, increasing specificity of the assay. Supplementing with a hybrid calibration method instead of customary peptide or concatamer calibration results in higher accuracy of PK analysis as well.
Application of Trapping Micro-LC–MS
T-µLC–MS with optimization enables distinct PK analysis of a drug and its effects on different tissues without the variation or chemical noise seen with other techniques. To demonstrate this significant method advantage, T-µLC–MS has been used to investigate the effectiveness of the drug adecetris (an ADC drug, Brentuximab Vedotin) in tumour penetration and biodistribution (4). Evaluation of the effects and safety of the drug through analysis of the plasma PK, tumour penetration, tissue distribution, drug-to-antibody ratio (DAR) value, and target antigen delivery validate the drug’s efficacy, carrier efficiency, and delivery method.
Optimized Tµ-LC–MS provides an integrated method to quantify the average DAR value, mAb concentration, and free monomethyl auristatin E (MMAE) toxin concentration in various tissues. The method validated the targeting effects of the drug by revealing a tumour concentration 10× higher than in other tissues, indicating successful targeted delivery and low biodistribution. The study also showed that the average DAR values within the tumour demonstrated extensive decay, suggesting a need for additional investigation into the pharmacodynamics of the drug (5).
Conclusion
Biopharmaceutical quantification involves a complicated set of processes to ensure investigation is valid and accurate. T-µLC–MS improves current methods to allow robust, sensitive, and selective assays. Ongoing development of the quantitation method can further increase resolution and specificity through additional processes, such as high resolution-selected reaction monitoring (H-SRM) with triple quadrupole MS for better selectivity and true simultaneous quantification. Development and optimization of methods to expand the scope of biotherapeutics quantitation using LC–MS enables a better understanding of the efficacy and toxicity these therapeutics can have.
References
- B. An, M. Zhang, R.W. Johnson, and J Qu, Anal. Chem.87(7), 4023–9 (2015).
- M. Qu, B. An, S. Shen, M. Zhang, X. Shen, X. Duan, J.P. Balthasar, and J Qu, Mass Spectrom. Rev.36(6), 734–754 (2017).
- B. An, M. Zhang, and J. Qu, Drug Metab. Dispos. 42(11), 1858–66 (2014).
- J. Qu et al., manuscript in preparation
- J. Qu et al., manuscript in preparation
Debadeep Bhattacharyya is the Sr. Manager of Product Marketing (Triple Quadrupole MS) for all businesses in the applied market division at Thermo Fisher Scientific Inc. With more than 12 years of experience working with mass spectrometers and other high-end analytical instruments, and also with customers across varied industries and academic institutions, Dr. Bhattacharyya applies his expertise and the experiences he has gained in working with customers and ensuring their scientific and business success. He also works closely with scientists, business development personnel, regulatory bodies, and key opinion leaders to address the needs of mass spectrometer (specifically triple quadrupole) users. Deb received his Ph.D. in chemistry and biochemistry from Emory University (Atlanta, Georgia, USA).
Bo An is a graduate student in Jun Qu’s laboratory.
Jun Qu is the group leader of the proteomics and pharmaceutical analysis laboratory of SUNY-Buffalo and a professor in the Department of Pharmaceutical Sciences. His research is focused on the study of clinical and pharmaceutical proteomics and pharmaceutical analysis using LC–MS-based strategies.
E-mail:debadeep.bhattacharyya@thermofisher.comWebsite:www.thermofisher.com

















Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).
Silvia Radenkovic on Building Connections in the Scientific Community
April 11th 2025In the second part of our conversation with Silvia Radenkovic, she shares insights into her involvement in scientific organizations and offers advice for young scientists looking to engage more in scientific organizations.
Regulatory Deadlines and Supply Chain Challenges Take Center Stage in Nitrosamine Discussion
April 10th 2025During an LCGC International peer exchange, Aloka Srinivasan, Mayank Bhanti, and Amber Burch discussed the regulatory deadlines and supply chain challenges that come with nitrosamine analysis.
Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).
Silvia Radenkovic on Building Connections in the Scientific Community
April 11th 2025In the second part of our conversation with Silvia Radenkovic, she shares insights into her involvement in scientific organizations and offers advice for young scientists looking to engage more in scientific organizations.
Regulatory Deadlines and Supply Chain Challenges Take Center Stage in Nitrosamine Discussion
April 10th 2025During an LCGC International peer exchange, Aloka Srinivasan, Mayank Bhanti, and Amber Burch discussed the regulatory deadlines and supply chain challenges that come with nitrosamine analysis.
