Algorithms and Databases: Unlocking Non-Targeted Screening of Small Molecules with Ambient Ionization Mass Spectrometry
Almost all sectors of analytical chemistry are finding applications for ambient ionization mass spectrometry (AI–MS) because of its ease of use, speed of analysis, and sensitivity. Although emphasis has been placed on developing new hardware that can help analyze unique samples across various applications, there has not been much innovation in the functionality of software tools and mass spectral libraries to support applications like non-targeted searching. In this article, we discuss new algorithms and libraries that have enabled non-targeted analysis of small molecules using AI–MS, as well as some of the key considerations and outstanding questions in the field.
From forensic science (1) to healthcare (2) to food analysis (3), ambient ionization mass spectrometry (AI–MS) continues to find new application areas (4). The appeal of AI–MS—which is listed in almost every article on the topic—includes minimal to no sample preparation, rapid analysis times, the ability to alter ionization chemistry on the fly, and desirable sensitivity. However, there is a one (major) drawback to AI–MS, which is that the resulting mass spectra can be difficult to interpret. Given the lack of chromatography, an AI–MS spectrum consists of ions generated from all compounds in the sample overlaid on top of one another.
For some applications, like olive oil authentication (5) or diabetes detection (6), it may be logical to consider an AI–MS mass spectrum as a fingerprint of the sample. In this case, a barrage of statistical tools (for example, principal component analysis [PCA], linear discriminant analysis [LDA], and machine learning) can be applied to the collections of spectra to group which samples are similar, identify the potential source of a sample, or answer another classification-related question. Other applications, like forensic seized drug analysis, require the analyst to understand the precise chemical makeup of a sample. In these applications, treating an AI–MS mass spectrum as a fingerprint and using classification tools is inadequate.
Non-Targeted Analysis with AI–MS
So how does one accomplish non-targeted analysis with AI–MS? Given that most mass spectral search tools are designed for chromatography approaches where the spectra of pure compounds are compared, we need to use new approaches. Recognizing that most AI–MS sources are soft ionization techniques, and therefore typically produce intact molecular ions (that is, protonated or deprotonated molecules), one approach is to identify individual peaks in the spectrum using simple lookup tables of compound m/z values. Although this approach can be fruitful, lookup tables can provide inconclusive results when several compounds share the same m/z (for example, isomeric or isobaric compounds). This limitation is exacerbated when using unit resolution instrumentation.
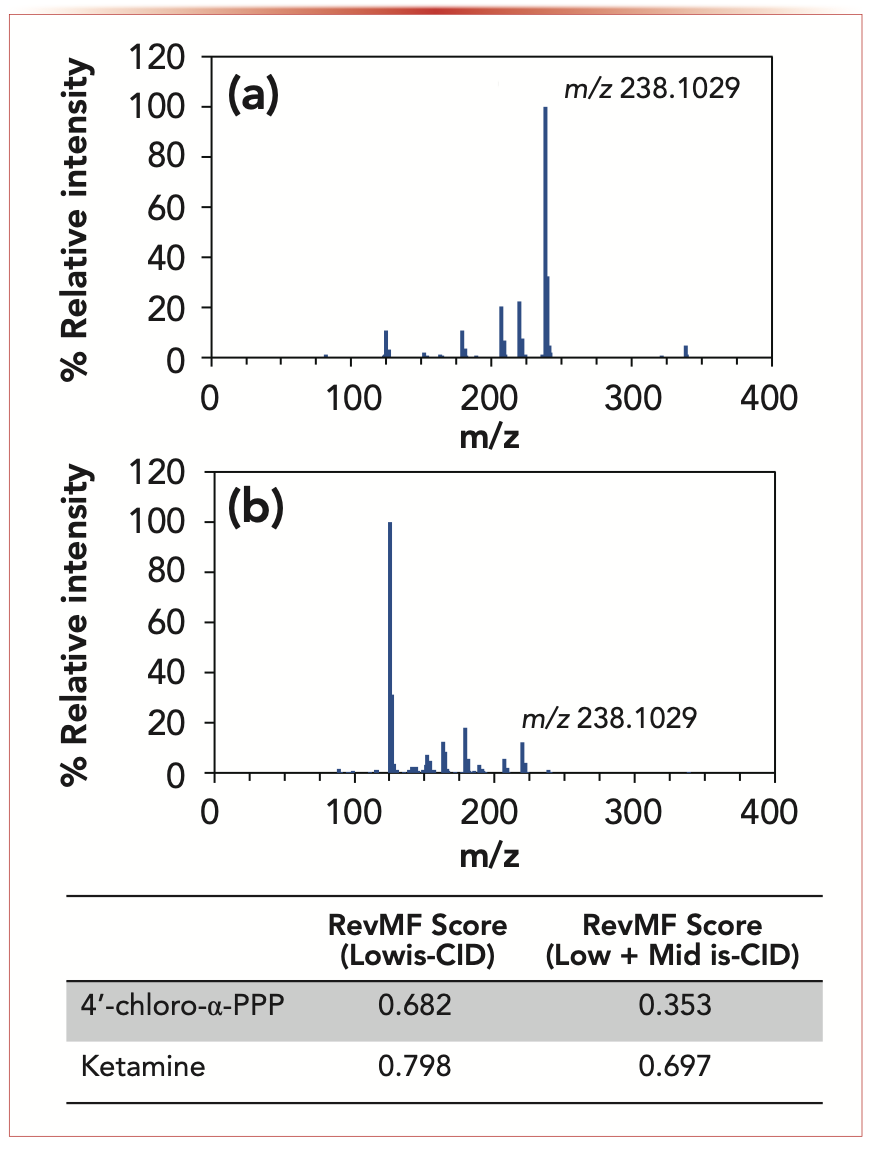
For example, take the AI–MS spectrum of a seized drug sample shown in Figure 1. The main peak at nominal m/z 238 could be attributed to ketamine, a common drug of abuse, 4’-chloro-α-pyrrolidinopropiophenone, an isomeric synthetic cathinone, or both. As a result, we need a different approach if we want to determine whether one or both compounds are truly in the sample.
FIGURE 1: An example of (a) a low energy is-CID mass spectrum and (b) a mid energy is-CID mass spectrum of a seized drug sample containing ketamine. The table at the bottom provides the reverse match factor (RevMF) scores for ketamine and 4’-chloro-α-pyrrolidinopropiophenone (PPP) obtained by analyzing the low and mid is-CID spectra with the ILSA. Note the significant drop in score for 4’-chloro-α-PPP when the mid is-CID spectrum is included in the search. Scores range from 0 to 1 with 1 being a perfect match.
There are currently two approaches that improve the specificity of the lookup table approach: tandem mass spectrometry (MS/MS) with collision induced dissociation (CID), or in-source CID (is-CID). In MS/ MS, we can specify that the m/z 238 ion should be isolated, fragmented, and a product ion scan collected. These product ions can then be used to help elucidate the structure of the ion. Of course, specifying an ion to be isolated is not ideal when performing a true non-targeted analysis, especially in AI–MS when your sample is only ionized for a few seconds. With is-CID, one or more “fragment spectra” are generated at multiple is-CID energies without any precursor ion isolation. The issue here is that product ions in fragmentation spectra cannot be directly associated with their precursors. If we rethink the data analysis approach, these high energy is-CID fragmentation spectra can actually be used as a rich source of information in non-targeted analysis.
Inverted Library Search Algorithm (ILSA)
The inverted library search algorithm (ILSA) is a new data analysis approach developed at the National Institute of Standards and Technology (NIST) to use multiple is-CID mass spectra and unlock non-targeted screening with AI–MS (7–9). As the name implies, the ILSA inverts the traditional mass spectral library search process. Instead of asking the traditional question of “Does my questioned spectrum match this known spectrum?”, we instead ask “Does this known spectrum match my questioned spectrum?” Given that AI–MS spectra are mixture spectra, there is an important additional piece to the ILSA question. Specifically, we ask, “Does the known spectra match a sub-pattern within my questioned spectrum?” The sub-pattern piece is critical because we must assume our questioned spectrum is not pure. Although we can do this on just one is-CID spectrum (a low energy is-CID spectrum that produces intact protonated ions), the value is increased when we combine multiple is-CID spectra, enabling us to not only match intact ions but also match fragment ions that are produced at higher energies. By implementing the ILSA alongside a similarity metric of choice, we can obtain scores that can be used for deciding the identity of a compound in the mixture. Consider our example in Figure 1. When utilizing the ILSA along with the reverse match factor and three is-CID spectra, we can see the scores from ketamine are much higher than those of 4’-chloro-α-pyrrolidinopropiophenone, allowing us to have more confidence that the sample contains ketamine.
To truly make use of any library search algorithm, one needs a high-quality mass spectral library. The ILSA currently uses the NIST Direct Analysis In Real Time MS (DART–MS) Foren- sics Database (10,11), a regularly updated resource that contains multiple is-CID mass spectra of over 1100 compounds of interest to the forensic community. The library was built and evaluated with the assistance of a combination of automated and interactive software tools that could be used to produce libraries of measurements collected with other AI–MS platforms as well.
Summary and Conclusion
There is still one major question that needs to be addressed before we can be fully certain about best practices for non-targeted analysis using AI–MS and the ILSA, and that has to do with the effectiveness of centrally collected spectral libraries. Given the rapidly changing landscape of AI sources and MS platforms, are the data collected using a particular source on a specific MS platform useful to a chemist using a different configuration? The corollary to that question is equally important—even using the same configuration of instruments, is there other laboratory-to-laboratory variability (12,13) that impacts the effectiveness of a centrally collected AI–MS spectral library? In traditional chromatographic techniques where ionization occurs under vacuum, spectral reproducibility across instruments and laboratories is quite high and so libraries (for example, the NIST series of electron ionization [EI]–MS libraries) are reliable resources. Some preliminary studies in our laboratory focused on the platform-to-platform utility of a central library are promising, but much more work needs to be done. Unfortunately, there is currently limited literature on the laboratory effects of AI–MS measurements (14–17). Answering these questions will help us determine the costs and benefits of centrally created libraries like the NIST DART- MS Forensics Database compared to using custom libraries created under the specific conditions of the laboratory.
Although our recent work has made non-targeted analysis with AI–MS practically possible, the final piece of the puzzle is the identification of novel compounds. The ILSA, like most mass spectral library search algorithms, is limited to identifying known compounds that are contained within the search library. For chromatographic techniques producing mass spectra of pure compounds, NIST has a few approaches (for example, MS Interpreter, hybrid search, fentanyl classifier) to assist in novel compound classification. These methods are not naturally amenable to is-CID mass spectra of mixtures but variations of them might be applicable with further work. Developing additional tools and resources for novel compound identification, using AI–MS or other mass spectral platforms, is a worthwhile pursuit.
Non-targeted screening using AI–MS and is-CID mass spectra could potentially provide critical, and previously unthinkable, capabilities to nearly all sectors of analytical chemistry in the future. Accomplishing this objective would require the development of algorithms and data analysis procedures that improve capabilities even more. Near-instantaneous non-targeted compound identification may seem far-fetched, but the research is moving in that direction and the road to success seems clear.
Disclaimer
Certain commercial products are identified to adequately specify the procedure. This does neither imply endorsement or recommendation by NIST nor does it imply that such products are necessarily the best available for the purpose.
These opinions, recommendations, findings, and conclusions do not necessarily reflect the views or policies of NIST or the U.S. Government.
References
(1) E. Sisco and T.P. Forbes, Forensic Chem. 22, 100294 (2021). https://doi.org/10.1016/j.forc.2020.100294
(2) H. Su, M.-Z. Huang, J. Shiea, and C.-W. Lee, Mass Spectrom. Rev. e21784 (2022). https://doi.org/10.1002/mas.21784
(3) A. Arrizabalaga-Larrañaga, J.F. Ayala-Cabrera, R. Seró, J.F. Santos, and E. Moyano, in Food Toxicology and Forensics, C.M. Galanakis, Ed. (Academic Press, Cambridge, 2021), pp. 271–312.
(4) S. Rankin-Turner and L.M. Heaney, Anal. Sci. Adv. 2(3–4), 193–212 (2021). https://doi.org/10.1002/ansa.202000135
(5) M. Beneito-Cambra et al., Trends Anal. Chem. 132, 116046 (2020). https://doi.org/10.1016/j.trac.2020.116046
(6) G. Zhang et al., Metabolomics 16(1), 11 (2020). https://doi.org/10.1007/s11306-020-1637-8
(7) A. Moorthy and E. Sisco, J. Am. Soc. Mass Spectrom. 32(7), 1724–1734 (2021). https://doi.org/10.1021/jasms.1c00097
(8) A.S. Moorthy, S.S. Tennyson, and E. Sisco, J. Am. Soc. Mass Spectrom. 33(7), 1260–1266 (2022). https://doi.org/10.1021/jasms.2c00090
(9) E. Sisco, A.S. Moorthy, S.S. Tennyson, and R. Corzo, J. Am. Soc. Mass Spec. 32(7), 1725–1734 (2021). https://doi.org/10.18434/MDS2-2448
(10) E. Sisco and A.S. Moorthy, NIST DART-MS Forensics Database (is-CID), (2020). https://doi.org/10.18434/mds2-2313
(11) E. Sisco, A.S. Moorthy, and L.M. Watt, J. Am. Soc. Mass Spectrom. 32(3), 685–689 (2021). https://doi.org/10.1021/jasms.0c00416
(12) B.J. McCullough and C.J. Hopley, Rapid Commun. Mass Spectrom. 35(S2), e8534 (2021). https://doi.org/10.1002/rcm.8534
(13) E. Gurdak et al., Anal. Chem. 86(19), 9603–9611 (2014). https://doi. org/10.1021/ac502075t
(14) G.A. Newsome, L.K. Ackerman, and K.J. Johnson, Anal. Chem. 86(24), 11977–11980 (2014). https://doi.org/10.1021/ac503652x
(15) S.R. Kumbhani, L.M. Wingen, V. Perraud, and B.J. Finlayson-Pitts, Rapid Commun. Mass Spec. 31(19), 1659–1668 (2017). https://doi.org/10.1002/rcm.7951
(16) C.L. Feider et al., J. Am. Soc. Mass Spectrom. 30(2), 376–380 (2019). https://doi.org/10.1007/s13361-018-2105-9
(17) G.A. Newsome, L.K. Ackerman, and K.J. Johnson, J. Am. Soc. Mass Spectrom. 27(1), 135–143 (2016). https://doi.org/10.1007/s13361-015-1259-y
Edward Sisco is with the National Institute of Standards and Technology, in Gaithersburg, MD. Direct correspondence to: edward.sisco@nist.gov

University of Tasmania Researchers Explore Haloacetic Acid Determiniation in Water with capLC–MS
April 29th 2025Haloacetic acid detection has become important when analyzing drinking and swimming pool water. University of Tasmania researchers have begun applying capillary liquid chromatography as a means of detecting these substances.
Prioritizing Non-Target Screening in LC–HRMS Environmental Sample Analysis
April 28th 2025When analyzing samples using liquid chromatography–high-resolution mass spectrometry, there are various ways the processes can be improved. Researchers created new methods for prioritizing these strategies.
New Study Reviews Chromatography Methods for Flavonoid Analysis
April 21st 2025Flavonoids are widely used metabolites that carry out various functions in different industries, such as food and cosmetics. Detecting, separating, and quantifying them in fruit species can be a complicated process.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)









