Advantages of Sequential or Bracketed Injection Methods to Improve the Chromatographic Analysis of Biotherapeutics
This article explores three case studies where autosampler injection programs help to (i) reduce the carry-over observed for large nucleic acids during anion-exchange chromatography (AEX), (ii) reduce peak distortion and breakthrough for monoclonal antibodies (mAbs) during hydrophilic-interaction chromatography (HILIC), and (iii) facilitate dissolution of lipid nanoparticles (LNPs) so that a size-exclusion chromatography (SEC) method can be applied for ribonucleic acid (RNA) payload analysis.
High-performance liquid chromatography (HPLC) analysis of biotherapeutics introduces new challenges because large molecules may require distinct injection modes or sample handling protocols to improve the analysis. Some challenges can be addressed by using autosampler methods that allow an analyst to program specific injection sequences. This article explores three case studies where autosampler injection programs help to (i) reduce the carry-over observed for large nucleic acids during anion-exchange chromatography (AEX), (ii) reduce peak distortion and breakthrough for monoclonal antibodies (mAbs) during hydrophilic-interaction chromatography (HILIC), and (iii) facilitate dissolution of lipid nanoparticles (LNPs) so that a size-exclusion chromatography (SEC) method can be applied for ribonucleic acid (RNA) payload analysis. The appropriate use of autosampler capabilities offers unique benefits to analyze biotherapeutics.
Modern autosamplers provide precise, accurate, and fast sample introduction into the mobile-phase flow path, and can also perform additional liquid handling steps such as sample dilution, mixing, internal standard addition, and precolumn derivatization (1,2). Significant savings in terms of reduced labor hours can be found in many different example projects an it is the dilution-related tasks that are most valuable.In addition to sample manipulation options, so-called “sequential injection” methods can also be applied. Sequential injections are most commonly used for two purposes. They can limit solvent mismatch problems, and they can also limit extra-column band broadening effects (3,4). A sequential injection method is often referred to as “bracketed injection” or “performance optimizing injection sequence” (POISe) when it is applied to improve band focusing.
The POISe technique involves injecting a defined volume of weak solvent (usually weaker than the mobile phase) along with the sample to increase solute retention during sample loading. This will result in a better match between the sample solvent and the mobile phase. Such a technique is particularly effective when gradient elution is applied, because it improves the peak shape and width of early eluting peaks. In isocratic elution, it can also reduce the detrimental effects of pre-column band broadening (especially when the solute retention is low) (5).
A number of investigators have previously reported the benefits of sequential injection methods (3–9). However, it is still rarely used in routine analysis.
The purpose of this article is to demonstrate the capabilities of sequential injection methods for the analysis of large molecule biotherapeutics. Three methods are discussed: (i) a recently introduced “salt plug injection method” to improve the carryover and recovery of messenger ribonucleic acids (mRNAs) in anion-exchange (AEX) separations, (ii) an “acetonitrile (MeCN) pre-plug injection method” to avoid peak deformation and breakthrough of monoclonal antibodies (mAbs) and related species in hydrophilic-interaction chromatography (HILIC), and (iii) an automated injection program to denature lipid nanoparticles (LNPs) so that their nucleic acid payloads can be analyzed by size-exclusion chromatography (SEC).
Salt Plug Injection Method for Improved AEX Analyses of Large Nucleic Acids
Although large-scale AEX separations (purification) are regularly performed for oligonucleotides and nucleic acids, very few analytical-scale AEX separation methods have been published. The reason for the limited use of AEX for analytical purposes is probably the lack of method robustness, poor recovery and high carryover effects, which are especially problematic for larger nucleic acids (10–12). Very long equilibration times between two sample injections may be required to reduce carryover (10). In addition to time-consuming equilibration steps, specific additives have also been used to suppress unwanted intermolecular interactions. Some cations have also been found to be beneficial because they stabilize the higher order structure of nucleic acids, and therefore improve recovery (12).
With the above considerations in mind, we recently proposed a new and efficient technique to significantly reduce carryover occurring in AEX separations (13). The phenomenon behind the high carryover effects observed with biomolecules (and macromolecules in general) is mostly related to non-desired secondary (or non-specific) interactions with surfaces and to both inter- and intramolecular interactions occurring in a macromolecular system.
Macromolecules are surface-active systems, which undergo non-specific adsorption when they come into contact with different types of surfaces (14). During an adsorption step, macromolecules undergo shape changes (that is, unfolding). The area on the adsorbent surface that is occupied by the macromolecules is often referred to as the “footprint” (15). The size of a footprint usually increases with the time available for adsorption. The increase of footprint size is sometimes referred to as “spreading.” If the solute concentration is high enough, the surface is occupied faster and therefore there is less time available for the spreading to occur. This results in a smaller average footprint. In addition to concentration and available time (residence time), other parameters such as solvent pH, ionic strength and temperature may also impact the spreading of the footprint and thus the recovery and carry-over observed in liquid chromatographic separations.
Nucleic acids in AEX follow a so-called on–off (bind and elute)-like elution mechanism (16). Therefore, it can be assumed that large nucleic acids, such as mRNAs, are bound at the head of the column and remain motionless until they experience the eluting mobile-phase composition. Hence, a certain time is available for the nucleic acid to spread and to establish multipoint interactions with the stationary phase. It is reasonable to assume that the binding interaction between nucleic acids and AEX ligands is inherently very strong (and increases as the residence time at the column inlet increases).
To limit the strength of the initial binding interaction, we propose that a program to inject the sample together with a strong solvent/eluent plug is created. To do this, we recently proposed bracketing nucleic acid samples with injection plugs containing a high concentration of salt (counterion). It is expected that the counterions—if injected in excess—will bind to the strong binding sites of the stationary phase and compete with the nucleic acid analytes.
If a large fraction of the stationary phase ligands is occupied by the counterions (at the time of the sample injection), then the nucleic acid spreading process will be inhibited and therefore a weaker binding interaction is expected to occur at the column inlet. In addition to high ionic strength, the pH of the solvent plug can also be adjusted to be close to the pKa of the stationary phase functional groups, that is, pH 10 in the case of a weak anion exchanger. The combined effect of salts and high pH is intended to limit the strength of the initial solute adsorption and therefore improve recovery and reduce carryover.
It was found that the effect of the salt plug injection method is sample dependent. Therefore, the following variables are worth studying and optimizing: (i) the volume of a salt pre-plug, (ii) the volume of a salt post-plug, (iii) the total volume of the bracketing salt plugs, (iv) the type of salt employed (NaCl, (NH4)2SO4, guanidine-HCl or NaBr are the most efficient salts) and (v) the pH of the salt plug. In general, the most effective injection procedure was found to be an injection sequence where the sample plug is bracketed with pre- and post-plugs of 2 M NaBr solution (at pH ~10).
Figure 1 shows a schematic view of a bracketed versus traditional injection. Please note that at pH higher than 10.5–11.0, nucleic acids (mRNAs) might be denatured, and the base-pairing and base-stacking interactions might be disrupted. At high pH, the intact RNA can thereby be linearized which can lead to significant spreading, a larger binding footprint, and stronger adsorption. Furthermore, some nucleobases can be deprotonated above pH 10, which would add additional negative charge to the nucleic acid analyte, and increase the retention time even further.
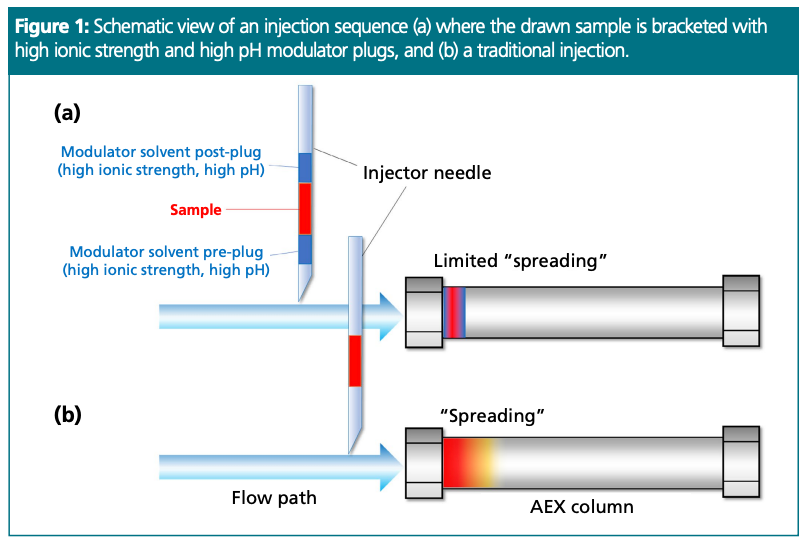
Based on our observations, carryover decreases, and recovery significantly improves with increasing ratio of modulator to sample volumes (Vm/Vs), until the ratio reaches a value of ~1.2–1.3. Above this “limit,” a fraction of the injected sample is taken by the salt plug and a partial breakthrough peak appears on the chromatogram. If value is greater than 2, then the entire mRNA peak is eluted at the column’s dead time (total breakthrough). When setting a (Vm/Vs) ≈1–1.2, carryover values as low as 1–3% can be reached instead of 8–20% which is often observed without salt plug modulation. Setting (Vm/Vs) ≈1 seems to be a good starting point. As an example, if a 2 µL volume of mRNA sample is going to be injected, then a good start is to program a sequence with a 1 µL modulator pre-plug + 2 µL sample + 1 µL modulator post-plug.
Figure 2 shows an example of a bracketed versus traditional injection for modified Cas9 mRNA (length: 4521 nucleotides). A weak AEX column was used with salt gradient elution. The 2 µL sample plug was bracketed with pre and post 1 µL 2 M NaBr (pH = 10.2) modulator solvent plugs. The red traces show the first blank injection after the sample. A carryover value of ~2% was observed with the bracketed injection, while carryover amounted to ~8–9% with the traditional injection.
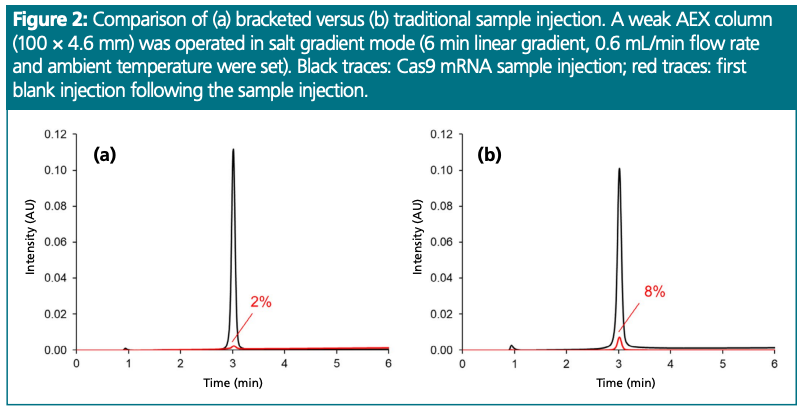
Our proposed approach helps to reduce the strength of solute (mRNA) binding at the head of the column and thus improves recovery and carryover in AEX. By using this bracketed injection method, the carryover of large mRNAs can be reduced to ~2%, in contrast to the 6–20% carryover which is often observed with conventional AEX methods.
Acetonitrile Plug Injection for Improved HILIC Analyses of mAbs and Therapeutic Proteins
When water-soluble biopharmaceuticals, such as mAbs, are analyzed in HILIC, there is an inherent mismatch between the sample diluent (aqueous) and the mobile phase, which is usually composed of a high proportion of an aprotic organic solvent such as acetonitrile (MeCN). A difference in eluent strength between the sample diluent and mobile phase can result in severe solute breakthrough and peak distortion effects. Adjustment of the sample diluent can minimize this mismatch, that is, increasing the ratio of MeCN in the sample diluent. However, this adjustment is very often impossible because of the nature of the sample. The use of diluents with a high proportion of MeCN might be counterproductive as denaturation phenomena or even precipitation might occur. Since the breakthrough effect is related to the injection volume, the term “critical injection volume” (Vcrit) is often used to express the maximum injectable volume that will not result in breakthrough (7,8). This critical volume directly depends on the column volume and the retention factor of the solute in the injection solvent (sample diluent). Due to the abovementioned on-off elution mechanism of large molecules, it can be expected that only a very low sample volume can be injected safely (typically Vcrit = 0.1–0.3 µL aqueous mAb sample into a 2.1 × 150 mm HILIC column).
To avoid the complications discussed above, we propose introducing aqueous mAb samples together with a MeCN solvent plug onto the column (17). Various injection programs were tested, (pre-, post- or bracketed injection plug). Interestingly, introducing the sample with a solvent pre-plug was found to be the most effective approach. Several sample to solvent plug ratios from 1:1 up to 1:10 were tested. Figure 3 shows a plot of “solvent to sample plug ratio” as a function of “sample plug volume to column volume ratio %”. The plot suggests that the required solvent plug volume depends on the sample volume injected. It has been found that the 1:10 ratio helps avoid breakthrough and is a valuable strategy for injecting relatively large volumes of therapeutic proteins, for example, mAb samples. As large as 2–5% of the column volume can be injected without any breakthrough effect. However, it should be noted that slight peak broadening may be observed when the sample to solvent plug ratio is high (greater than 5). Therefore, if a smaller sample volume injection is sufficient, the solvent/sample plug ratio can be reduced. Various flow rates were also investigated, and it was found that flow rate had no effect on the required solvent to sample plug volume ratio.
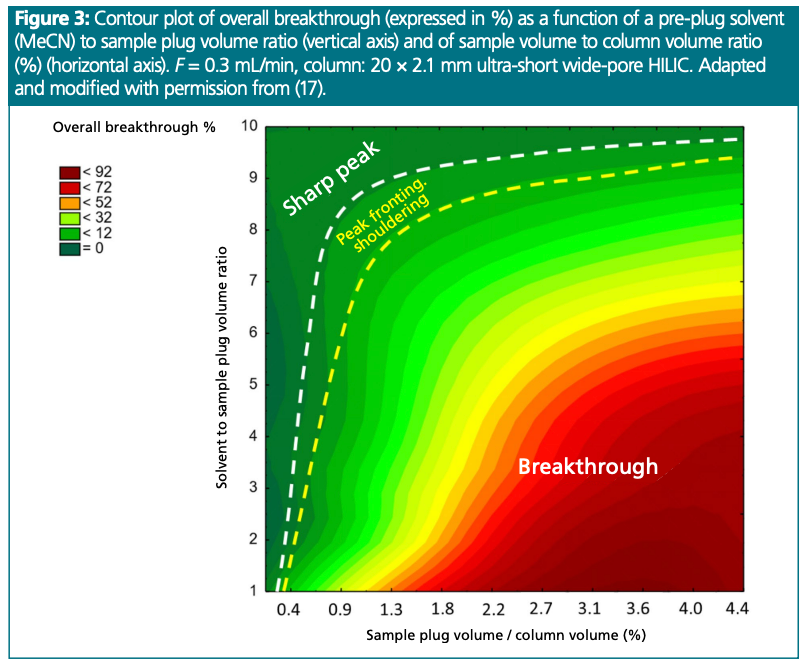
This strategy has already been applied to a wide range of therapeutic mAb products of different physicochemical properties (17). In all cases, relatively large volumes could be successfully injected onto small-volume HILIC columns (ultra-short columns) using purely aqueous samples. Note that this approach is not limited to protein injections; the same rules apply to other macromolecules, such as nucleic acids.
Programmed Injection Method To Analyze Nucleic Acids Released From Denatured LNPs
Large molecule biotherapeutics can also benefit from programmed injection methods to facilitate sample preparation. Apart from increased throughput and labor-savings, automation is also beneficial to handle sensitive or hazardous samples, for example infectious viral vectors, reducing the possibility of sample handling artifacts and exposure to hazards. There is a growing body of evidence that autosampler liquid handling functions can be successfully applied to more complex biopharmaceuticals products, as it was used to prepare dilutions of adeno-associated virus (AAV) samples that resulted in highly accurate calibration curves (18).
Nucleic acid-loaded LNPs are an example of highly delicate species that can show changed physical properties upon experiencing relatively low mechanical stress (19). For this reason, we investigated the potential of programmed injections to prepare sample dilutions for SEC separations that are becoming increasingly common for such samples (Figure 4) (20).
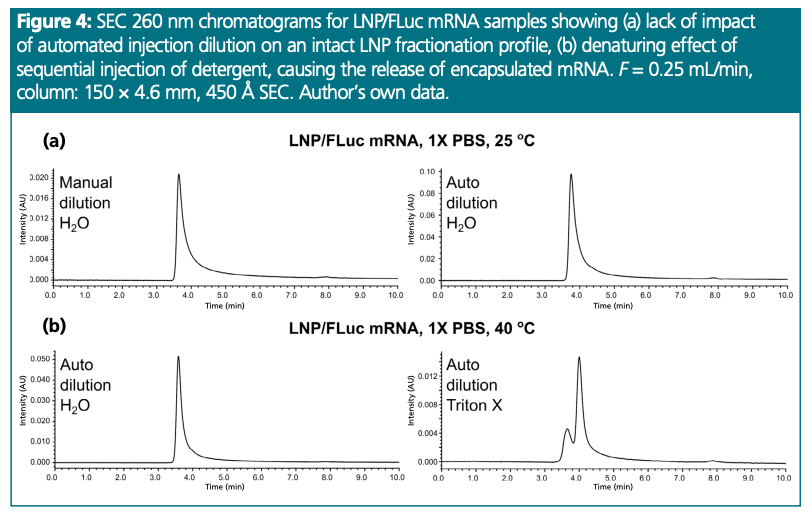
Firstly, we compared an SEC profile obtained from a direct injection of manually water-diluted intact LNPs (1 μL of 0.05 mg/mL loaded with FLuc mRNA) or auto-sampler diluted solutions (5 μL of 0.1 mg/mL and 5 μL of water) (Figure 4[a]). The applied non- denaturing conditions led to comparable fractionation of both samples, indicating the suitability of this method for handling of LNPs.
Moreover, to show that such solutions can be efficiently manipulated, the LNP sample was diluted with a detergent—Triton X-100 Detergent (Figure 4[b]) which is known to cause deformulation of the lipid nanoparticles and the release of their payload (Note: Triton X-100 Detergent itself is a hazardous chemical, because it is a phenol-based surfactant. Its impurities and degradants can act as endocrine disruptors, and exposure to analysts and the environment should be minimized. Alternatively, the use of a more ecologically friendly non-ionic surfactant, such as Brij 58 Detergent, can be explored.). In this work, Triton X-100 Detergent was indeed applied, and an injection of LNP/FLuc mRNA (1 μL of 0.1 mg/mL) sample was combined with a plug of aqueous Triton X-100 solution (1 μL of 0.4% v/v). This resulted in the disruption of the particle and allowed the detection of the encapsulated nucleic acid, which was observed as a peak eluting at a later elution time. This non-optimized experiment indicated that the denaturing action of the surfactant can be achieved. This is a proof-of-concept experiment; as shown in Figure 4(b) it can be seen that the extent of denaturation of the LNP is incomplete. Further optimization of the technique might be possible by adjusting the number of mixing cycles, injection delay, and volume/concentration ratios.
Conclusion
Programmed (multi-step or multi-vial) injections are most commonly used for sample preparation (dilution, internal standard addition, mixing, derivatization). However, sequential (plug, bracketed) injection methods may offer more advantages than initially realized. These types of injection modes can be applied to improve recovery and reduce breakthrough effects related to mismatched diluent to initial condition mobile phase compositions. It is large molecule analytes that can benefit most from sequenced injections.
In AEX, the recovery of mRNA can be significantly improved by performing a bracketed injection. By introducing the sample between pre- and post- plugs of high pH salt solution (that is, 2 M NaBr at pH ~10), carryover values can be reduced to 1–3%. Without bracketed injections, carryover of intact mRNAs can be as high as 10–20%.
In HILIC, injecting the sample along with an acetonitrile pre-plug resulted in symmetrical and sharp peaks. No breakthrough was observed. When applying a relatively large solvent plug volume (that is, 5–10 times larger than the sample volume), the overall loaded sample volume can be significantly increased. As much as 2–5% of the column volume can be injected without any breakthrough effect.
In SEC, LNP samples can be efficiently manipulated via automated sample preparation. This allows development of protocols for automated dilutions or disruptions of the intact LNP sample, potentially increasing reproducibility of the results and reducing impact of human induced errors.
Overall, we aimed to show that programmed injections can not only facilitate biotherapeutics sample preparation, but they can also be advantageously applied to address new types of chromatographic challenges.
Acknowledgement
The authors would like to acknowledge Christian Reidy (Waters Corporation) for reviewing the manuscript.
Declaration of Competing Interest
The authors are employees of Waters (Milford, MA, USA), a manufacturer of chromatography systems and consumables. Cas9 mRNA and FLuc mRNA were purchased from TriLink BioTechnologies. The Fluc mRNA/LNP was purchased from PackGene Biotech – Cap 1+N1meψUTP+LNP (Firefly Luciferase-mRNA-SM-102). Triton is a trademark of Union Carbide Corporation. Brij is a trademark of Croda Americas LLC.
References
(1) Steiner, F.; Paul, C.; Dong, M. W. HPLC Autosamplers: Perspectives, Principles, and Practices. LCGC North Am. 2019, 37 (8), 514–529.
(2) Kataoka, H. Sample Preparation for Liquid Chromatography. In Liquid Chromatography: Applications, 3rd ed.; Handbooks in Separation Science, Vol. 2; Fanali, S., Haddad, P. R., Riekkola, M.- L., Chankvetadze, B., Poole, C. F., Eds.; Elsevier, 2023; pp 1–48. DOI: 10.1016/B978-0-323-99969-4.00006-1
(3) Vanderlinden, K.; Broeckhoven, K.; Vanderheyden, Y.; Desmet, G. Effect of Pre- and Post-Column Band Broadening on the Performance of High-Speed Chromatography Columns Under Isocratic and Gradient Conditions. J. Chromatogr. A. 2016, 1442, 73–82. DOI: 10.1016/j.chroma.2016.03.016
(4) Anspach, J. A.; Sanchez, C. A.; Farkas, T. Injection Sequence for Optimizing Performance in UHPLC Separations. Phenomenex, 2011. http://phx.phenomenex.com/lib/po89900911_L_2.pdf (accessed 09-25-2023).
(5) Sanchez, A. C., Anspach, J. A.; Farkas, T. Performance Optimizing Injection Sequence for Minimizing Injection Band Broadening Contributions in High Efficiency Liquid Chromatographic Separations. J. Chromatogr. A. 2012, 1228,338–348. DOI: 10.1016/j.chroma.2012.01.038
(6) Taylor, M. R.; Kawakami, J.; McCalley, D. V. Managing Sample Introduction Problems in Hydrophilic Interaction Liquid Chromatography. J. Chromatogr. A. 2023,1700, 464006. DOI: 10.1016/j.chroma.2023.464006
(7) Chapel, S.; Rouvière, F.; Peppermans, V.; Desmet, G.; Heinisch, S. A Comprehensive Study on the Phenomenon of Total Breakthrough in Liquid Chromatography. J. Chromatogr. A. 2021, 1653, 462399. DOI: 10.1016/j.chroma.2021.462399
(8) Chapel, S.; Rouvière, F.; Heinisch, S. Comparison of Existing Strategies for Keeping Symmetrical Peaks in On-Line Hydrophilic Interaction Liquid Chromatography x Reversed-Phase Liquid Chromatography Despite Solvent Strength Mismatch. J. Chromatogr. A. 2021, 1642, 462001. DOI: 10.1016/j.chroma.2021.462001
(9) Gritti, F.; Sanchez, C. A; Farkas, T.; Guiochon, G. Achieving the Full Performance of Highly Efficient Columns by Optimizing Conventional Benchmark High-Performance Lquid Chromatography Instruments. J. Chromatogr. A 2010, 1217, 3000–3012. DOI: 10.1016/j.chroma.2010.02.044
(10) Bridonneau, P.; Bunch, S.; Tengler, R.; et al. Purification of a Highly Modified RNA-Aptamer: Effect of Complete Denaturation During Chromatography on Product Recovery and Specific Activity. J. Chromatogr. B: Biomed. Sci. Appl. 1999, 726 (1–2), 237–247. DOI: 10.1016/S0378-4347(99)00037-7
(11) Ferreira, G. N. M.; Monteiro, G. A.; Prazeres, D. M. F.; Cabral, J. M. S. Downstream Processing of Plasmid DNA for Gene Therapy and DNA Vaccine Applications. Trends Biotechnol. 2000, 18 (9), 380–388. DOI: 10.1016/s0167-7799(00)01475-x
(12) Murphy, J. C.; Wibbenmeyer, J. A.; Fox, G. E.; Willson R.C. Purification of Plasmid DNA Using Selective Precipitation by Compactation Agents. Nat. Biotechnol. 1999, 17, 822–823. DOI: 10.1038/11777
(13) Fekete, S.; Lauber, M. Salt Plug Injection Methods for Improved Anion Exchange Analyses of Large Nucleic Acids; Application Note 720007991; Waters Corporation: Milford, MA, 2023. https://www.waters.com/nextgen/us/en/library/application-notes/2023/salt-plug-injection-methods-for-improved-anion-exchange-analyses-of-large-nucleic-acids.html (accessed 2024-06-13).
(14) Höger, K.; Becherer, T.; Qiang, W.; Haag, R.; Friess, W.; Küchler, S. Polyglycerol Coatings of Glass Vials for Protein Resistance. Eur. J. Pharm. Biopharm. 2013, 85 (3A), 756–764. DOI: 10.1016/j.ejpb.2013.04.005
(15) Rodriguez-Aller, M., Cusumano, A.; Beck, A.; Guillarme, D.; Fekete, S. Importance of Vial Shape and Type on the Reproducibility of Size Exclusion Chromatography Measurement of Monoclonal Antibodies. J. Chromatogr. B 2016, 1032, 131–138. DOI: 10.1016/j.jchromb.2016.04.032
(16) Fekete, S.; Yang, H.; Wyndham, K.; Lauber, M. Salt Gradient and Ion-Pair Mediated Anion Exchange of Intact Messenger Ribonucleic Acids. J. Chromatogr. Open 2022, 2, 100031. DOI: 10.1016/j.jcoa.2022.100031
(17) Pérez-Robles, R.; Fekete, S.; Kormány, R.; Navas, N.; Guillarme, D. Improved Sample Introduction Approach in Hydrophilic Interaction Liquid Chromatography to Avoid Breakthrough of Proteins. J. Chromatogr. A 2024, 1713, 464498. DOI: 10.1016/j.chroma.2023.464498
(18) Imiołek, M.; Fekete, S.; Kizekai, L.; Addepalli, B.; Lauber, M. Fast and Efficient Size Exclusion Chromatography of Adeno Associated Viral Vectors with 2.5 Micrometer Particle Low Adsorption Columns. J. Chromatogr. A, 2024, 1714, 464587. DOI: 10.1016/j.chroma.2023.464587
(19) Kamiya, M.; Matsumoto, M.; Yamashita, K.; et al. Stability Study of mRNA-Lipid Nanoparticles Exposed to Various Conditions Based on the Evaluation Between Physicochemical Properties and Their Relation with Protein Expression Ability. Pharmaceutics 2022, 14 (11), 2357. DOI: 10.3390/pharmaceutics14112357
(20) D’Atri, V.; Imiołek, M.; Quinn, C.; Finny, A.; Lauber, M.; Fekete, S.; Guillarme, D. Size Exclusion Chromatography of Biopharmaceutical Products: From Current Practices for Proteins to Emerging Trends for Viral Vectors, Nucleic Acids and Lipid Nanoparticles. J. Chromatogr. A 2024, 1722, 464862. DOI: 10.1016/j.chroma.2024.464862
E-mail: Matthew_Lauber@waters.com
Website: www.waters.com

Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).













