A Stoichiometric Displacement Model for Proteins in Reversed-Phase LC
The literature on reversed-phase liquid chromatography (LC) of proteins is reasonably well developed, but not taught in the college classroom much. Kevin Schug therefore wants to focus on a stoichiometric displacement model for reversed-phase LC of proteins and why it is particularly insightful from a practical perspective.
Photo Credit: LAGUNA DESIGN/Getty Images
Kevin A. Schug, University of Texas Arlington, Texas, USA.
The literature on reversed-phase liquid chromatography (LC) of proteins is reasonably well developed, but not taught in the college classroom much. Kevin Schug therefore wants to focus on a stoichiometric displacement model for reversed-phase LC of proteins and why it is particularly insightful from a practical perspective.
As one might expect, intact protein separations are significantly more complex than small-molecule separations. Yet, with the current spotlight on biomarker discovery and quantitation, as well as biotherapeutic protein drug development, the need to handle and perform chromatographic separations on intact proteins is constantly increasing. In our group, we have been thinking a lot about this lately, and we are just starting to put some of our initial studies into the literature. One in particular that I am quite proud of is our recent demonstration of multiple reaction monitoring on a triple-quadrupole mass spectrometer as a means to detect intact proteins, with no digestion necessary.1 Now that we can detect our targets, we can turn our attention to high-efficiency separations. The literature is reasonably well developed on reversed-phase liquid chromatography (LC) of proteins, but I do not think that this is something that gets taught in the college classroom to a significant degree.2 Here I would like to focus on a paper from Geng and Regnier3 that describes a stoichiometric displacement model for reversed-phase LC of proteins, one that I found to be particularly insightful from a practical standpoint.
The authors describe retention of proteins on reversed-phase chromatography supports as a function of the number of solvent molecules (Z) needed to displace an adsorbed protein and cause its elution. Of course, the nature of the protein (relative hydrophobicity and hydrophilicity), the nature of the solvent (including bulk organic concentration and the type and concentration of modifiers), and the nature of the chromatographic stationary phase (density of alkyl ligands) all influence the process. Yet, for a given mobile- and stationary-phase system, each protein will have a unique value of Z, which can be used to characterize its retention and elution behaviour, relative to other proteins in that system.
There are a few aspects about proteins and their chromatographic behaviour that are worth noting here. Some of these are also key to basic assumptions made in construction of the model. The concept that proteins will interact with the surface through multiple amino acid residues may seem obvious, but this concept is important. Multiple interactions can cause mixed-mode retention effects, but this phenomenon is not covered extensively in the article. The next is that proteins exhibit a very steep binding isotherm. In other words, the window between infinite retention and nonâretention for a protein is generally only a few percent change of organic solvent in the mobile phase. This property does appear to be correlated with the size of
the protein, with the window getting smaller (the isotherm getting steeper) as proteins get larger. Practically, this is why you cannot use isocratic methods to effectively separate a mixture of proteins. In the end, the model assumes that the protein displaces some solvent from the stationary phase when it adsorbs. The protein is then desorbed when the solvating power of the mobile phase becomes strong enough to displace the protein. In other words, the solvent must be able to break the multiple contact points between the protein and the stationary phase. The authors acknowledge that proteins have 3D structure and can change conformation, even while adsorbed to the chromatographic media. In fact, the authors do an exceptional job of explaining all of the various considerations regarding the plausibility and the limitations of the model in this article. It is very easy to read and understand.
Once all of the equilibria between a protein, a solvent, and a support material are defined, a relationship between capacity factor k’ and displacing agent concentration [D0] can be made:
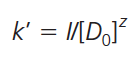
[1]
where I is a constant (for a given protein in a given system) that includes the phase ratio of the chromatographic system and a composite equilibrium constant. For experimental testing, this equation is better represented in logarithmic form:

[2]
where Z is the slope in a plot of log k’ versus log 1/[D0]. The parameters Z and log I determined in this manner can be tabulated and compared for proteins as average diagnostics of reversed-phase LC retention for different experimental conditions.
Under predominantly denaturing conditions (that is, 50–60% formic acid with isopropanol as displacing agent), such plots are nicely linear. For a series of seven model proteins ranging in molecular weight from 3335 to 44,000 g/mol, the number of solvent molecules displaced by protein adsorption (Z) was quite different for each and ranged from 2.59 to 23.8 (relative errors in these determined values were 0.2 to 0.5%). Z correlated quite well with molecular weight under these conditions, except in some cases where it was conjectured that the protein of interest was not fully dissociated. Because a fully dissociated protein could be expected to present a higher number of hydrophobic contact points with the reversed-phase LC support surface relative to a non-denatured form, it is easy to imagine that partial folding would alter the expected trend.
Further discussion is given to the effect of different solvents. For example, isopropanol is a rather strong displacing agent, so if a weaker solvent is used, then Z (as well as I) increase. When one looks at the eluotropic strengths of solvents, methanol (ε0 = 1.0) < acetonitrile (ε0 = 3.1) < isopropanol (ε0 = 8.3) < n-propanol (ε0 = 10.1). A nice discussion of the effects of different solvents, as well as other variables in protein separations, can be found in a more recent work by Dillon and colleagues.4 In any regard, isopropanol is more than twofold stronger than acetonitrile, perhaps the most commonly encountered reversedâphase LC mobile phase component for protein separations today, as an elution solvent for proteins. Yet, it should take significantly more acetonitrile molecules to displace a protein from a reversed-phase LC support compared to isopropanol, according to the stoichiometric displacement model. It should be noted that acetonitrile was not considered in the article, and my understanding is that this choice was made because it behaves quite differently in terms of its solvation of alkyl ligands on the chromatographic support than the series of alcohols, which were considered. In other words, assumptions were made to simplify the model based on similarities between homologous alcohols; the situation may be somewhat different for acetonitrile. Through a cursory search, I did not find any articles specifically investigating this point.
The Geng and Regnier article also describes the effects of common acidic pairing agents, such as phosphoric acid, formic acid, and trifluoroacetic acid, on protein retention. In general, phosphoric acid and formic acid are considered to be hydrophilic ion-pair reagents that will make proteins more soluble. In contrast, trifluoroacetic acid is a hydrophobic ionâpair reagent; its ion pairs are generally more strongly retained in reversed-phase LC. The incorporation of trifluoroacetic acid is also common to improve peak shape. Even though phosphoric acid is considered to solvate proteins well, Z values with equivalent concentrations of trifluoroacetic acid are equal to or lower than those obtained with phosphoric acid. Further, small changes in trifluoroacetic acid concentration (for example, 0.1% versus 0.3%) can have drastic and variable effects on protein retention; Z values generally decrease further with higher concentrations of trifluoroacetic acid, but not for all proteins. This decrease is believed to occur because of the amphiphilic nature of trifluoracetate. At lower concentrations, it may effectively ion pair with cationic sites (and shield interactions with residual silanols). At higher concentrations, it may also pair with hydrophobic regions of the protein.
I believe that this stoichiometric displacement model is a very nice instructional tool to help visualize and practically treat protein retention in reversed-phase LC. Many later works have built on this model. As noted by the authors, and agreed upon here, the situation is certainly much more complex than a couple of defining values. Trends can be established, but there appear to often be exceptions to those trends. These are most likely a result of the changing 3D structure of a protein in a reversedâphase LC system, or the difficulty to predict whether a protein will resist denaturing or not, under certain chromatographic conditions. Even so, if one wants to develop reversed-phase LC systems that provide altered selectivity, then many of the concepts described by this model can be leveraged. At the very least, I hope that this post prompts readers to look back into the earlier literature on this topic - there is a lot of good information, and we should not reinvent the wheel.
References
- E.H. Wang, P.C. Combe, and K.A. Schug, J. Am. Soc. Mass Spectrom. (2016). doi:10. 1007/ s13361-016-1368-2
- K.A. Schug, “Intact Protein Separations: Some Education Is Missing”, The LCGC Blog. 13 November 2014. http://www.chromatographyonline.com/lcgc/Blog/The-LCGC-Blog-Intact-Protein-Separations-Some-Educ/ArticleStandard/Article/detail/860052.
- X. Geng and F.E. Regnier, J. Chromatogr.296, 15–30 (1984).
- T.M. Dillon, P.V. Bondarenko, D.S. Rehder, G.D. Pipes, G.R. Kleeman, and M.S Ricci, J. Chromatogr. A1120, 112–120 (2006).
Kevin A. Schug is a Full Professor and Shimadzu Distinguished Professor of Analytical Chemistry in the Department of Chemistry & Biochemistry at The University of Texas (UT) at Arlington. He joined the faculty at UT Arlington in 2005 after completing a Ph.D. in Chemistry at Virginia Tech under the direction of Prof. Harold M. McNair and a post-doctoral fellowship at the University of Vienna under Prof. Wolfgang Lindner. Research in the Schug group spans fundamental and applied areas of separation science and mass spectrometry. Schug was named the LCGCEmerging Leader in Chromatography in 2009 and the 2012 American Chemical Society Division of Analytical Chemistry Young Investigator in Separation Science. He is a fellow of both the U.T. Arlington and U.T. System-Wide Academies of Distinguished Teachers.

Characterizing Plant Polysaccharides Using Size-Exclusion Chromatography
April 4th 2025With green chemistry becoming more standardized, Leena Pitkänen of Aalto University analyzed how useful size-exclusion chromatography (SEC) and asymmetric flow field-flow fractionation (AF4) could be in characterizing plant polysaccharides.
Investigating the Protective Effects of Frankincense Oil on Wound Healing with GC–MS
April 2nd 2025Frankincense essential oil is known for its anti-inflammatory, antioxidant, and therapeutic properties. A recent study investigated the protective effects of the oil in an excision wound model in rats, focusing on oxidative stress reduction, inflammatory cytokine modulation, and caspase-3 regulation; chemical composition of the oil was analyzed using gas chromatography–mass spectrometry (GC–MS).












