Open Access, Automation, and the Promise of Simple Complexity
Columnist Michael Balogh looks at the questions: Where are we now? What do the early MS innovators see as their legacy? And what can we expect in the future?
The oxymoron in the title illustrates a predicament in analytical practice. Since the early 1990s, we have witnessed great strides in mass spectrometry (MS), improvements that bring sophisticated capabilities to a diverse user base. Yet along with this success has come the inevitable learning process. So where are we now? What do the early innovators see as their legacy, and what can we expect according to those who now have 15 or 20 years of experience behind them?

Michael P. Balogh
Pharmaceutical practice was the leading edge of a movement allowing a fully trained mass spectrometrist to give nonspecialists access to advanced capability without the need for extensive training. To it we owe such terms of art as "open access" and "walk-up MS," which characterize a distributed rather than centralized analytical capability. The belief underpinning this distributive capability was that medicinal chemists and others could perform high-order analytical processes without the need for extensive training on rather complicated instruments. It is not automation, or a machine performing repetitive tasks with little or no human intervention that frees chemists to pursue more creative activities or those requiring discriminating judgment. Instead, open access and walk-up systems provide a menu of processes from which chemists presenting a variety of samples choose analytical tasks that run automatically.
A wide range of automated laboratory practices has evolved to include all industries. Symyx Technologies, Inc. (Santa Clara, California), which first developed automation to advance its own research, is an example of a company that grew with the opportunity and necessity. Its initial market objectives were not drug discovery but material science programs: for example, producing petrochemical and polymer catalysts that led to high-profile collaborations with ExxonMobil and Dow. The company's expertise in polymer chemistry led to founding and then spinning off Ilypsa Pharmaceuticals, recently acquired by Amgen (Thousand Oaks, California). Though research remains a significant part of Symyx's operations, it nevertheless enjoys a reputation as a third-party integrator of laboratory workflows, with major pharmaceutical customers as well giants in other industries to its credit (www.symyx.com).
While I was researching this topic, a colleague made an interesting point. Cecilia Mazza (Senior Market Development Manager, Chemical Analysis, Waters Corporation, Milford, Massachusetts) noted that the terminology we use to describe aspects of workflow capability remains fluid as its adoption and integration evolves. She observes that it is sometimes difficult to ascertain the extent of a workflow when terms like "on-line," "at-line," and "off-line," which we blithely assume to be standard, become blurred with terms such as "near-line."
Open access began with the tools available at the time (gas chromatography [GC] and solid analysis probes) to suit a need (increasing throughput while decreasing repetitive task necessities for humans). As laboratory technology advanced, bringing with it ever wider use of the mass spectrometer, and as the nature of compounds investigated became increasingly diverse, the need persisted.
Open Access: In the Beginning
The idea to investigate this subject began with a discussion with one of the first people I met in MS practice, Jeffrey P. Kiplinger, who was then at Pfizer and is now president of Averica Discovery Services (Worcester, Massachusetts). Kiplinger, one of the early adopters and developers of the open-access concept, collaborated with software engineers at Fisons Instruments (Manchester, UK) division that was renamed VG Biotech in 1995 and, later, Micromass. Waters purchased Micromass in 1997 and continued the company's manufacture of mass spectrometers.
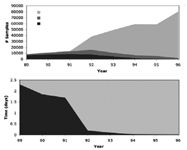
Figure 1: Effect of introducing Open Access MS to the discovery chemistry environment at Pfizer in the early 1990s. Shown are plots of samples analyzed per year by technique (top) and the mean time required for analysis per year (bottom). (Taken from an internal Pfizer presentation courtesy of Jeffrey Kiplinger.)
"As an organic chemist with a technology bent, I backed into a mass spectrometry career. From postdoc work with Maurice Bursey and Alan Marshall, I took my first real job in 1988, with Pfizer, to run a mass spec service group for a discovery chemistry department of about 75 chemists. Pfizer was then the world's 20th largest pharmaceutical company — before my interview I had to learn how to spell 'Pfizer' and find an annual report in the library to learn what the company made."
"At Pfizer, my job was to set up a new capability, FAB-MS [fast-atom bombardment], which promised the capability of acquiring spectra on many of the new chemical leads the company was working on — compounds with higher molecular mass and more polar functionality than a lot of common small-molecule drugs. While the new high-resolution, double-focusing magnetic sector instrument was being ordered and awaiting installation, I received a one-year education in how mass spec contributed to the work of drug discovery."
"My associate Dick Ware, universally known as 'Tupper,' and I acquired spectra on tiny samples of final synthesis products intended for registration in the compound file. These were purified compounds of known structure — the chemists had NMR spectra of every intermediate compound on the synthesis path, as well as the final product — and we were providing the final verification. We dissolved the samples and, using a fine syringe, added a microliter to the end of a capillary tube. Then we used tweezers to place the tube at the end of a long, cannon-like, direct-insertion probe. The probe went into the mass spec's ion source through two vacuum locks, and after a pump-down [evacuation] cycle, we heated the probe and boiled off the sample, acquiring perhaps as many as a half-dozen +EI spectra. A little more than half of those spectra showed a molecular ion; the remaining spectra (with no significant molecular ion present) gave fragment ions that we hand-annotated with structures (this was where our knowledge of McLafferty's and Turecek's Interpretation of Mass Spectra was important). With an external calibration, we could achieve mass accuracy to ±10 ppm on about 80% of the structures, so the chemist performing the synthesis could publish or patent."
"As mind-numbing as this work sounds — 12 spectra a day, little creativity, no productive activity one day each month because of source cleaning — it was fun learning how drug discovery worked, sharing the load with really smart scientists, and working in a well-funded lab. Many of the chemical series we worked on gave similar spectra, and over time we learned the characteristic fragment ions these series produced. FAB ionization gave us a "molecular ion" [MH+] much more frequently, and the chemists much preferred it to the older electron ionization [EI] data. When I asked several of them why, their answer surprised me:
'We really only look at the molecular ion. If the ion is wrong, it means we did something wrong. If it's right, it's confirmation that the product is the right stuff.'
"Apparently, all the extra work, spectral annotation and mass measurement, were needed less than mass confirmation. Over a few weeks, it became apparent that as little as one in 50 submissions required careful inspection by the "trained mass spectrometrists." The rest were fully useful if they showed the expected molecular mass peak."

Figure 2: AIDE automation of the NMR process workflow at Pfizer. (Courtesy of Wei Wang, presented at Association of Laboratory Automation 2004, San Jose, California.)
"As a postdoc at Ohio State University, I published some work jointly with Professor Leo Pacquette's group. Dr. Pacquette was then the best-funded organic chemist in the department. His large group even had its own mass spec, a Hewlett-Packard GC–MS system that everyone in the lab used without the help of a mass spectrometrist. One grad student kept the instrument operational, and everyone simply put samples in the open autosampler tray and typed the entry into a sample list. They used a short capillary column and a rapid temperature gradient. The system proved indispensable for checking the purity of solvents and starting materials and even produced usable spectral data on a variety of products. When the student chemists couldn't confirm identity with GC–MS, they resorted to NMR analysis. At the time, my biggest lesson was that a mass spectrometrist was not always necessary in obtaining analytical data. Many of my colleagues didn't learn that lesson until much later."
"Back at Pfizer, by the summer of 1990 we were running as many as 1000 samples per month [using an initial 'open access' GC system], and chemists were reporting useful structural data on a surprising number of compounds. We ordered a second system. Other than an initial 30-minute training session and regular maintenance, this GC–MS system ran without intervention by analytics staff. We never saw the data generated unless asked a question, and so to my knowledge, that instrument was the first open-access mass spectrometer (we called it "walk up") in the pharmaceutical industry."
"In 1991 I had some discussions with a Pfizer counterpart from the U.K. Research division in Sandwich, Frank Pullen. We were encouraged by the success of the walk-up GC–MS experiment, and wondered whether the newer liquid interfaces such as thermospray (TSP) and particle beam (PB) might offer a way to acquire spectra on compounds that were too large or thermally labile to pass through a GC column. By 1992, a Fisons Trio-1000 TSP-MS system in the U.K. was operational along with an HP-5989 with a PB–ammonia CI interface in the U.S. Both gave molecular mass peaks on 80%–90% of discovery chemists' compounds. Flow injection analyses were performed on a one-minute cycle. No ionization method then seemed stable enough with liquid chromatography (LC) flows or gradients to give smooth data over a complete LC run, and the 20-minute high performance liquid chromatography (HPLC) gradients of the early 1990s were too long to implement on a walk-up system."
"The instruments were custom built, and they included innovations unavailable on commercial production instruments. Such modifications are incorporated in today's instruments. But at the time, they were six-to-nine-month development projects for the manufacturers, and they represented some cost to Pfizer. They included a software interface for easy sample login, an ability to add samples to a running queue, and an automated idle or shutdown routine so that an unattended instrument didn't continue to run. No one had anticipated an unattended mass spectrometer, and no one had previously thought of controlling an LC system from the MS data system."
"At the 1993 ASMS meeting in San Francisco, these two instrument system approaches were presented at a workshop on automated MS technology. At the time, these instruments, having each run 40,000 to 60,000 samples over the preceding year, were probably the most productive individual mass spectrometers ever built. The notion that unsophisticated users could operate mass spectrometers and that spectra could be returned directly to a chemist without interpretation by a mass spectrometrist was regarded as risky and establishing a poor precedent. Some members of the audience were clearly threatened. I wrote down one quotation, which I won't attribute:
"Putting mass spectrometry in the hands of the end-user is simply irresponsible!"
"When we published our experiences with this new approach in early 1995, I titled the paper 'Putting Mass Spectrometry in the Hands of the End User' (1). Next to our paper in the Journal of the American Society of Mass Spectrometry was a similar experiential account by Lester Taylor, Bob Johnson, and Robert Raso that first used the term 'open access' to describe a similar approach at what was then Burroughs-Welcome Pharmaceuticals (now GSK) (2)."
Kiplinger's perspective is instructive: "Neither Hewlett-Packard nor the Fisons division that built the Trio-1000 MS were particularly interested in commercializing a walk-up platform. But VG Biotech picked up the project, trademarked the 'open access' phrase and ran with it. I was told that in 1996 the open access platform generated over $30 million in revenue, an unheard-of annual production for a mass spectrometer."
"By the mid-1990s the drug discovery world was in the thrall of technology and investing resources at an unprecedented rate. High-throughput technologies for screening, chemistry, and analysis consumed so much of the industry's attention that for a time, it seemed companies forgot to use them to discover drugs. The pressures on chemists to produce and on technologists and analysts to enable production were huge, and they generated some memorable conflicts. Mass spectrometrists told stories of users who sabotaged each other by removing queued samples and substituting their own. Analysts told of users running black, tarry samples or vials half-filled with silica from scraped thin-layer chromatography (TLC) plates (clogging systems). Some groups named and shamed abusers on a 'wall of shame.' Others simply gritted their teeth and hired more support staff to keep systems operating."
"Meetings in the 1995–2000 era focused on high-throughput technologies, now mostly linked to LC–MS. We debated the proper ratio of supporting analytics staff to chemists to run an effective discovery program. Technologies for remote instrument monitoring were presented that allowed us to see our machines from home, to check them on the weekends. For a time it was a hot role to play in pharma — developing new productivity enhancement tools that would ultimately increase the ability to discover new drugs by 100-fold or more."
Of course, not all of that happened, while remote monitoring is commonplace now. "Bubbles pop, and we've returned to a saner way of thinking." Nevertheless, the value to a chemist of being able to acquire useful information is a constant. "Throughout the history of organic chemistry, understanding what happens in a reaction has always been a bottleneck. The little leap of understanding that took place in about 1990 was that the molecular mass of the product is the clearest and simplest indicator of the success of a synthesis. The development of open-access LC–MS, a work of nearly 10 years of partnership between pharma customers and technology companies, was driven by the value inherent in this realization. It's hard to imagine a modern chemistry program without an available open-access mass spec system."
Open Access: Today
For the most part, Kiplinger has moved on to new challenges while today's practitioners improve on the benefit of open access. What do they have to say about it? In the following sections, we will look into where the open-access concept is headed, taking into consideration economics, technological advances, and — to the extent we can — the vicissitude of human nature.
In March 2009, with the help of William Farrell, Senior Principal Scientist in High Throughput Analysis and Purification, Pfizer Global Research (La Jolla, California) and a director for the Society of Small Molecule Science, which organizes the CoSMoS conference each year (www.CoSMoScience.org), we assembled a roundtable composed of people with scientific understanding and extensive practical knowledge in the area of open access. John Van Antwerp, recognized for his role in developing and implementing open access in laboratories throughout the Americas for Waters Corporation joined in the discussion with a group of leaders in the practice of distributed high end technology.
Farrell has more than 20 years chromatographic experience, specializing in supercritical fluid chromatography (SFC) and HPLC and hyphenated MS techniques. Since 1998 he has been working on implementing SFC and HPLC for the analysis and purification of combinatorially derived medicinal compounds. He has been a major driving force behind the development of automated preparative SFC systems and has pioneered the use of SFC for high throughput applications. We wanted to address the question of whether the instruments and their software allow seamless agreement between need and function. John Isbell, Ph.D, Director of Analytical Sciences and DMPK, Genomics Institute of the Novartis Research Foundation (GNF), La Jolla, California, worked for 10 years in mass-directed purification, including a couple of years in the Kassel group during which time he worked on a custom, parallel, two-channel PrepLCMS that was ultimately made obsolete by the MUX Purification Factory (Waters Corporation). He moved to GNF to set up a HT-purification group around the MUX Purification Factory and demonstrated its ability to purify 10,000 samples/month. John Harman, Associate Director, Research, Neurocrine Biosciences, Inc. La Jolla, California, has been leading efforts since 1997 in mass directed purification starting with open access followed by open access by mass directed purification until 2006 when the model switched to centralized or service purification. Muhammad Alimuddin , Senior Scientist, Pfizer Global R&D, La Jolla involved with the Discovery Platform Chemistry/Open Access has been administering open access systems for more than 9 years. Initial experience was in open access administration of LC, LC–MS, and GC–MS in a CRO laboratory. At Pfizer he expanded the open access portfolio by introducing reversed phase mass directed prep purification, chemical stability assay and photo stability assay using LC–MS, assessing residual solvents by GC–MS and high resolution accurate mass using an LC–time-of-flight (TOF) system. Wei Wang, Ph.D, Associate Research Fellow, Pfizer Global Research, La Jolla, California, started as an NMR spectroscopist at Princeton University in 1995, and spent the last 10 years applying his skills of physical and analytical chemistry, as well as analytical informatics at Pfizer. Wang brought complementary insights to the discussion with his experience with automating open access NMR. His current interests include two different platforms of high-throughput, capillary flow or capillary tube NMR on Varian and Bruker.
The group determined that, in some cases, the need that first gave rise to open access persists. In pharmaceutical practice, because analytical support is primarily for medicinal chemists, and mass spectrometers involve significant costs in both initial outlay and training, their use must be warranted. Medicinal chemists are well trained in NMR practice as part of their schooling and, as Wei Wang pointed out, open access or walkup NMR offers the same advantages of open access LC–MS, namely, greater access to expensive instruments, faster turn around time and reduced overall costs.
NMR lends itself to open access development through inherent simplicity and cooperative human behaviors. Factors like cost and complexity may keep LC and MS out of the hands of many during their school years. As Farrell points out,
"I'm sure there are very progressive academic labs but . . . the vast majority . . . know how to run [manual] columns, TLC, and how to perform NMR analyses. Other technology is foreign to [students]. If every NMR were converted to flow-probe NMR, you would run into an equal number of problems as in HPLC."
Wei Wang adds "Tube-based NMR tends to have fewer problems but there is still room to improve its robustness and ease of use."
Wang and colleagues at Pfizer developed a walk-up workflow for NMR, with a collection of designed user interfaces and utility programs that incorporated third-party vendors' processes into a customized process. Their goal was to use automation to increase productivity by reducing manual, repetitive tasks, increasing reliability, decreasing response and down times, and simplifying user interfaces to help users obtain better information faster. They derived an acronym, "AIDE" from the exercise:
- Automate everything we can.
- Integrate various software and technologies into one "turn-key" solution.
- Develop our own process to meet users' needs.
- Educate users to utilize our facility effectively.
Often, despite training coupled with experience, the most expedient approach is to centralize the practice, as Neurocine's John Harman discovered. He found open-access practice for purification generally inefficient. When deciding how to proceed, he and his team ignored instrument usage time because "the cost of a machine is nothing compared to, unfortunately, the intangible cost of time." After analyzing runs for a year, he found chemists were successful in purifying their molecules only about 40% of the time. Yet according to the chromatography, they should have been successful 70% of the time. Thus, the chemists were essentially throwing away 30% of their samples simply because they were not preparing them properly, choosing the wrong gradient, or setting the wrong threshold.
The consequence was to centralize the purification practice. Harman identified super users and trained them for 6 h. He observed that users compete with each other and used the new knowledge to their advantage. When someone needs assistance they come to the central group for help. Lack of training in analytical chemistry leads to the incidents Kiplinger alluded to, which earned certain individuals their membership on the "wall of shame."
John Isbell at GNF uses a different approach to ensure purification remains a distributed capability in open access. GNF has "medicinal chemists who police their own instruments." Isbell relates, "We never asked the chemists to do this. What actually happened is that the experienced chemists took it upon themselves to enforce department policies, reminding others what constitutes proper usage. The medicinal chemists made all the rules and policies for all the walk-up instruments. A key element in the success of this approach is attributed to the very low turnover at GNF."
Other steps have proven useful, too. A company with two sites approached work submissions differently. One group took the approach that the job of chemists was to make compounds, not run LC–MS. So although the chemists worked without constraint, their administrator spent 30% of the time unclogging injectors from submitters using sample solutions made at "20 mg/mL in 90% water."
The twin site had as many systems but required using only the vials supplied to submit samples. They supplied 4-L bottles of methanol and trained the operators simply "if you had enough material to perform NMR on your sample dilute it by 1000 add one drop of water and filter to check for solubility issues before doing LC–MS."
The difference between the groups was a 70% up-time at the first site and a 99% up-time percentage at the second. A significant difference underscoring the approach, according to our experts, rests on the size of a company. Peer pressure works at a smaller company with limited turnover, but a larger, more diverse company might benefit from common-sense training.
Open Access: Tomorrow
Particularly for small groups, solubility and other assays can be automated where those tasks are sufficiently repetitive and well-characterized. For example, a group can develop a chiral separation using extensive automation specifically for the purpose of creating a menu choice for open-access submitters. Thus, whereas chemists in contract research organizations are adept at using each piece of laboratory equipment, switching as needed from one project to another, those working in major pharmaceutical operations are being driven toward automation as a way to make more efficient using their time.
Muhammad lists various new techniques developed as open access: assessing residual solvents by GC–MS; high resolution, accurate mass using an LC–TOF instrument; reversed-phase, mass-directed, preparative purification (5–1000 mg crude loading); chemical stability assay using LC–MS; photo stability assay using LC–MS. He predicts "there is demand and we may expand more in providing physical properties of compounds like Log D, pKa, HLM based on a paper published last year . . . we are exploring the possibility of determine the Log D in flow mode to make it open access"(3). A thorough view of postcombinatorial thinking as we look forward to automated processes in pharmaceutical applications is presented in a comprehensive article by John Isbell (4).
Michael P. Balogh "MS — The Practical Art" Editor Michael P. Balogh is principal scientist, MS technology development, at Waters Corp. (Milford, Massachusetts); a former adjunct professorand visiting scientist at Roger Williams University (Bristol, Rhode Island); cofounder and current president of the Society for Small Molecule Science (CoSMoS) and a member of LCGC's editorial advisory board.
References
(1) F.S. Pullen, G.L. Perkins, K.I. Burton, R.S. Ware, M.S. Teague, and J.P. Kiplinger, J. Am. Soc. Mass Spectrom.6, 394–399 (1995).
(2) L.C.E. Taylor, R.L. Johnson, and R. Raso, J. Am. Soc. Mass Spectrom. 6, 387–393 (1995).
(3) M. Alimuddin, D. Grant, D. Bulloch, N. Lee, M. Peacock, and R. Dahl, J. Med. Chem. 51(16), 5140–5142 (2008).
(4) J. Isbell, J. Comb. Chem.10, 150–157 (2008).

Accelerating Monoclonal Antibody Quality Control: The Role of LC–MS in Upstream Bioprocessing
This study highlights the promising potential of LC–MS as a powerful tool for mAb quality control within the context of upstream processing.
Using GC-MS to Measure Improvement Efforts to TNT-Contaminated Soil
April 29th 2025Researchers developing a plant microbial consortium that can repair in-situ high concentration TNT (1434 mg/kg) contaminated soil, as well as overcome the limitations of previous studies that only focused on simulated pollution, used untargeted metabolone gas chromatography-mass spectrometry (GC-MS) to measure their success.
Prioritizing Non-Target Screening in LC–HRMS Environmental Sample Analysis
April 28th 2025When analyzing samples using liquid chromatography–high-resolution mass spectrometry, there are various ways the processes can be improved. Researchers created new methods for prioritizing these strategies.
Potential Obstacles in Chromatographic Analyses Distinguishing Marijuana from Hemp
April 28th 2025LCGC International's April series for National Cannabis Awareness Month concludes with a discussion with Walter B. Wilson from the National Institute of Standard and Technology’s (NIST’s) Chemical Sciences Division regarding recent research his team conducted investigating chromatographic interferences that can potentially inflate the levels of Δ9-THC in Cannabis sativa plant samples, and possible solutions to avoid this problem.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)









