Molecular Imprinting for Sample Preparation
LCGC North America
Molecularly imprinted polymers can replace conventional sorbent materials in sample preparation techniques such as solid-phase extraction (SPE), solid-phase microextraction (SMPE), and matrix solid-phase dispersion (MSPD), offering increased selectivity over the target analytes.
Sample preparation is the most crucial step for the development of an analytical method. The main purpose of sample preparation is the extraction and preconcentration of the target analytes, as well as the removal of the matrix interferences, before their separation and determination. It is the most time consuming step that should be deliberately optimized to enhance extraction selectivity and detection capability. Liquid–liquid extraction (LLE) and solid-phase extraction (SPE), along with their variations, are usually applied for sample extraction and cleanup. Molecularly imprinted polymers (MIPs) can replace conventional sorbent materials in sample preparation techniques such as SPE, solid-phase microextraction (SMPE), and matrix solid-phase dispersion (MSPD), offering increased selectivity over the target analytes. Attention is given in molecularly imprinted SPE (MISPE), which is mostly employed to study MIP applications, as well as a commercially available technique.
Molecularly imprinted polymer (MIP) synthesis involves the copolymerization of a functional monomer and a crosslinker around a selected template molecule, and, after template removal, a polymeric three-dimensional material with imprinted cavities is obtained. These cavities are complementary to the template molecule, and act as rebinding sites for the template or selective binding sites for analytes with analogous structure and functional group layout (Figure 1) (1).
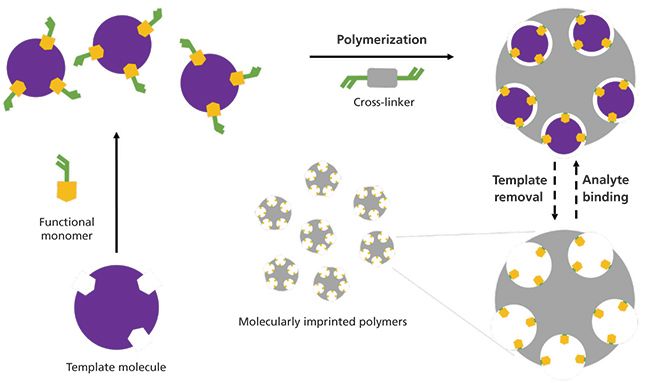
Figure 1: Molecular imprinting schematic representation. The template molecule assembles with the functional monomers and polymerize with the cross-linker. The template is removed from the developed polymers to free the imprinted cavities.
Molecular imprinting approaches rely on the interactions between the template molecule and the functional monomer, which can be either covalent or noncovalent. In covalent imprinting, covalent bonds are formed between the template and the monomer during MIP synthesis, as well as between the binding sites and the target molecule during MIP application. It is usually referred to as stoichiometric imprinting, and the developed MIPs display homogeneous cavity distribution and minimized incomplete binding sites, thus exhibiting improved selectivity. However, there are limited reactions that form covalent bonds between the template and the functional monomer that are reversible under mild conditions, resulting in slow analyte binding and unbinding.
Noncovalent imprinting involves the development of noncovalent interactions, such as hydrogen bonding, van der Waals forces, π-π and hydrophobic interactions, electrostatic forces, and metal coordination, both in MIP synthesis and binding–unbinding. Noncovalent imprinting is mostly preferred for MIP preparation, because of its simplicity, versatility of the compounds that can be imprinted, and the variety of commercially available functional monomers. However, the formation of a stable template–functional monomer complex requires relatively high amounts of the functional monomer, which usually leads to heterogeneously distributed and incomplete binding sites in comparison with a covalent approach.
Semi-covalent imprinting is a combination of the covalent and noncovalent imprinting, in which the template–monomer complex is formed by covalent interactions and analyte binding by noncovalent interactions. The semi-covalent approach offers stable template–functional monomer complexes during the imprinting process, and fast interaction between the target analyte and the binding sites. However, there are limited template–monomer combinations that meet this condition (2–5).
The critical factor in MIP synthesis is the selection of the template molecule, the functional monomer, and the cross-linker. The selected template should be chemically stable during the polymerization reaction, and contain functional groups that can interact with the monomer and not inhibit polymerization. Templates are mainly the target analytes. The selected monomer should interact with the template covalently or noncovalently to form a stable template–monomer complex, thus affecting the selectivity and binding capacity of the binding sites. Typical monomers include 4-vinylpyridine (4-VP), acrylic acid (AA), acrylamide (AM), methacrylic acid (MAA), 2-hydroxyethyl methacrylate (HEMA) and trifluoromethyl acrylic acid (TFMMA), with MAA being one of the most commonly used monomers in MIP synthesis. The selected cross-linker should copolymerize with the template–monomer complex to form a durable highly cross-linked polymer. Apart from the type, the quantity of the cross-linker used during MIP synthesis is crucial. Low quantity results in low cross-linked polymers with unstable imprinted cavities, while high quantity results in reduced number of imprinted cavities. Common cross-linkers include ethylene glycol dimethacrylate (EGDMA), divinylbenzene (DVB) and trimethylpropane trimethacrylate (TRIM) (1).
Apart from the effect of each individual factor in the MIP synthesis, the template–functional monomer/cross-linker ratio should be optimized to increase the MIP quality and selectivity. Optimization can be performed by top-down approaches, such as the trial-and-error approach based in which polymers with different ratios are tested until MIPs with the desirable selectivity are acquired. Another approach involves the optimization of the MIP synthesis in small scale for subsequent up-scaling. On the other hand, bottom-up approaches involve molecular level chemical interaction studies such as nuclear magnetic resonance (NMR), IR, and UV–vis spectroscopies and molecular modeling (2,4).
Template Removal
After the polymerization is complete, template removal is required. In this step, the template molecule used for the MIP synthesis is removed to free the imprinted cavities and enable analyte rebinding. Template removal can be considered more important than optimized MIP synthesis; however, in many cases, not much detail is given about optimizing this step. Insufficient template removal decreases the available imprinted cavities, leading to reduced MIP extraction efficiency, and template bleeding can occur in analytical applications, resulting in errors. Attempts for sufficient template removal may sometimes require extreme extraction conditions (pH, temperature, and MIP swelling) that can result in cavity breakage during template removal, cavity breakdown after template removal, or binding site deformation (6).
Template removal can be achieved by solvent extraction and physically assisted extraction. Solvent extraction includes MIP incubation in organic solvents, Soxhlet extraction, and the use of supercritical carbon dioxide and subcritical water. Physically assisted extraction includes pressurized liquid extraction (PLE), ultrasound-assisted extraction, and microwave-assisted extraction. Complete template removal is difficult, because of inability of the solvent to reach the most cross-linked regions of the MIPs and break the interactions between the cavities and the template. The most commonly used techniques are Soxhlet extraction and incubation in organic solvents. In Soxhlet extraction, the MIPs are washed over and over with a heated organic solvent that promotes template removal. A Soxhlet apparatus is affordable, easily operated, and common in many laboratories, and the process is applicable for any polymer composition and does not require a filtration step after the extraction. On the other hand, it is a rather time-consuming procedure (6–24 h) with increased organic solvent requirements (50–300 mL), and the solvents reach high temperatures that can cause template and MIP structure degradation. Incubation of the MIPs in organic solvents favors template extraction under milder conditions, thus preventing the disadvantages of Soxhlet extraction. However, it is a time-consuming procedure; also, increasing the solvent volume, heating, and stirring can accelerate the extraction process (6).
Non-Imprinted Polymers (NIPs)
Parallel with MIP synthesis, non-imprinted polymers (NIPs) are prepared by following the same synthetic protocol without the use of a template molecule. Therefore, NIPs do not possess imprinted cavities (Figure 2). NIPs act as a "control" to evaluate the strength of the interactions between the synthesized MIPs and the template that are specific in the case of MIPs, and nonspecific for NIPs. For this reason, binding tests are conducted by introducing the same amounts of MIPs and NIPs to solutions of predetermined concentration of the template molecule and measuring the bound concentration. The adsorption isotherms obtained from the binding tests and the use of Langmuir–Freundlich models can provide information about the number of the imprinted cavities, and their selectivity between the template and analogue molecules. Furthermore, MIPs and NIPs are tested in terms of particle size, shape, porosity, and surface composition (7).
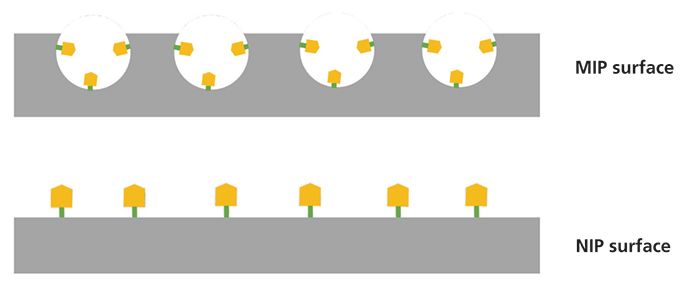
Figure 2: MIP and NIP surface schematic comparison. MIP surface imprinted cavities interact specifically with the analytes, and the NIP surface interacts nonspecifically.
Polymerization Techniques
MIP particles used in SPE protocols (8) can be prepared by bulk, suspension, emulsion, and precipitation polymerization. Bulk polymerization is a noncovalent approach for MIP synthesis, and is the most commonly used technique for fast development of sorbents for MIP solid-phase sample extractions. Bulk polymerization results in MIP monoliths that should be crushed, grounded, sieved, and sedimented with a proper solvent to receive MIP particles of the desirable size. However, this procedure results in particles with low extraction capacity, because of their varying size and shape, irregular distribution, or even complete lack of binding sites. Additionally, it requires large amounts of the template molecule with potential template leakage. In suspension polymerization, droplets of the polymerization mixture are suspended in a liquid phase, such as water, perfluorocarbon, or mineral oil. However, the resulting MIP particle size is not uniform. In emulsion polymerization, the template, the functional monomer, and the cross-linker are emulsified in an aqueous phase, creating an oil–water biphasic system; a surfactant is added to prevent diffusion. However, the surfactant sometimes interacts with the binding sites, thus reducing the binding capacity of the developed MIPs. In precipitation polymerization, the template is mixed with a solution of the functional monomer and the cross-linker in excess of the porogen solvent, and, as polymerization reaction occurs, the developed MIP particles precipitate (2,4,5).
Surface imprinting is a solid-phase imprinting approach in which a MIP layer is formed onto different substrates. Specifically, in core–shell imprinting, the MIP size, distribution, and nanoparticle functionality can be easily managed. The large surface area of the core particles increases the accessibility of the analytes to the MIPs, and faster analyte binding kinetics. Aggregation of imprinted nanoparticles can be minimized by adding a shell layer, or by surface modification. Cores used for the preparation of the imprinted particles can be Fe3O4 and SiO2 particles, quantum dots, or other polymers that bestow the MIPs with further properties. Magnetic core–shell MIP nanoparticles are an example of core–shell imprinting (4,5)
MIPs in Solid-Phase Extraction
Conventional SPE sorbents do not provide high selectivity over specific analytes, and sometimes matrix compounds are retained by the sorbent and eluted with the target analytes, thus causing problems during the analysis. MIPs can be employed as SPE sorbents to overcome these problems. Following the typical SPE procedure, molecularly imprinted solid-phase extraction (MISPE) cartridges are prepared by packing up to 500 mg of the prepared MIPs into polyethylene cartridges. The cartridges are then preconditioned, loaded with the sample solution, washed to remove the nonspecifically bound interferences, and the analytes are eluted with an appropriate solvent (Figure 3). The sample is usually prepared in a low-polarity solvent, so that the interactions with the binding sites are the highest, and analyte elution is carried out by a solvent that can break these interactions, and free the bound analytes (4).
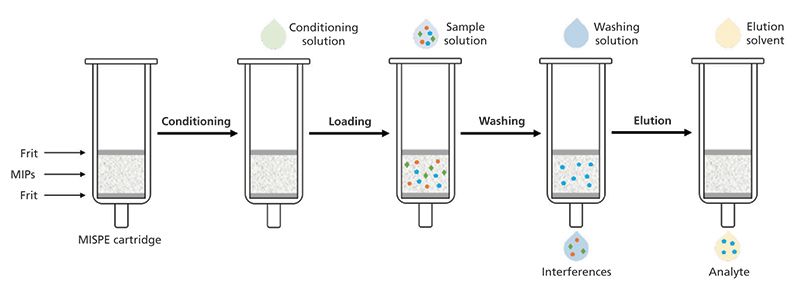
Figure 3: MISPE protocol schematic representation. The MIPs are packed into the SPE cartridge and preconditioned, the sample solution is then loaded, interferences are removed with a washing solution, and the analyte is eluted with an elution solvent.
Magnetic solid-phase extraction (MSPE) eliminates the packing step of the sorbent material into the SPE cartridge. The magnetic particles can be handled with the application of a magnetic field; thus, the extraction procedure is simplified and is less time-consuming. An amount of the magnetic particles is dispersed in the sample solution, the analytes are extracted and the particles are collected and separated from the solution by means of an external magnet. Analyte elution is carried out by repeating the same process with elution solvent (Figure 4a). Magnetic MIP nanoparticles can be prepared by core-shell imprinting that involves the coating of the surface of Fe3O4 particles with a functionalized SiO2 layer prior to polymerization reaction (Figure 4b). In comparison with other MIP particles, magnetic MIPs exhibit increased surface area with well distributed and accessible imprinted cavities and specific binding selectivity. However, their synthesis is tedious (9).
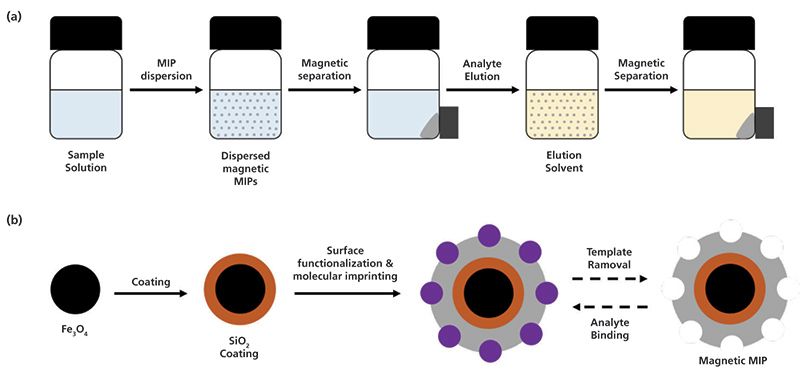
Figure 4: Schematic representation of (a) MSPE protocol with magnetic MIPs and (b) core-shell imprinting for the preparation of magnetic MIPs.
MIP Drawbacks
MIPs are of great interest due to reduced synthetic cost, increased chemical and physical stability, and MIP reusability. On the other hand, molecular imprinting is time-consuming, and complications such as heterogeneous imprinting can occur, whereas macromolecule imprinting is complicated. In the case of sample preparation, template bleeding and incompatibility with polar solvents can restrict MIP applications. Insufficient template removal results in template release during the elution step, which is called template bleeding and is a severe complication in the case of trace analysis. Thorough template removal (see the "Template Removal" section), along with thermal annealing, can reduce the template bleeding effect. Furthermore, the use of a structural analogue of the template molecule (also known as "dummy" template), during MIP synthesis, semicovalent imprinting, and surface imprinting are promising solutions. In many cases, analyte extraction must be performed in aqueous sample solutions. Most MIPs are prepared in organic solvents, but polar solvents inhibit the interactions between the template and the monomer, and reduce MIP recognition capability. Hydrophilic monomers and water as porogenic solvent can be used for the preparation of water compatible MIPs. A two-step extraction procedure that involves the extraction of the analytes from the aqueous phase to an organic solvent and then MIP extraction can also be employed (10).
MIP Applications
Further details for molecular imprinting (11) and MIP applications in SPE and other sample preparation techniques can be found in the literature (12–15). MIPs have been reviewed as selective sorbents in bioanalysis (16) for the determination of macromolecules (17), drug analysis (3,9) for the determination of nonsteroidal inflammatory drugs (18), and food analysis (5) for the determination of antibiotics in milk samples (19). Furthermore, MIPs were applied for the determination of personal care products (20), ochratoxin A (21,22), drugs of abuse (23), and the extraction of organophosphorus pesticides (7), agrochemicals (24), and contaminants and pollutants in environmental samples (4,25).
Commercially Available MIPs
There is a significant number of published papers that have described individual protocols for the synthesis and application of MIPs for the extraction of various analytes from different sample type. MISPE cartridges for the extraction of antibiotics (chloramphenicol, fluoroquinolones, aminoglycosides, and nitroimidazoles), pharmaceuticals (clenbuterol, beta-agonists, beta-blockers, and nonsteroidal inflammatory drugs), polyaromatic hydrocarbons, bisphenol A, nitrosamines, patulin, and riboflavin are commercially available.
Conclusion
Overall, there is no denying that MIPs can be successfully applied for the extraction and determination of various analytes, offering increased selectivity and reusability. Although there are some commercially available MISPE formats, there is a lot of work to be done so that MIPs can be prepared and applied on a daily basis for routine analysis in analytical laboratories. Future perspectives lean toward improvements and innovation for MIP synthesis, template removal, and MIP application format.
References
(1) L. Chen, X. Wang, W. Lu, X. Wu, and J. Li, Chem. Soc. Rev. 45, 2137–2211 (2016). doi:10.1039/C6CS00061D.
(2) E.N. Ndunda and B. Mizaikoff, Analyst 141, 3141–3156 (2016). doi:10.1039/C6AN00293E.
(3) S. Ansari and M. Karimi, Talanta 164, 612–625 (2017). doi:10.1016/j.talanta.2016.11.007.
(4) A. Speltini, A. Scalabrini, F. Maraschi, M. Sturini, and A. Profumo, Anal. Chim. Acta 974, 1–26 (2017). doi:10.1016/j.aca.2017.04.042.
(5) J. Ashley, M.A. Shahbazi, K. Kant, V.A. Chidambara, A. Wolff, D.D. Bang, and Y. Sun, Biosens. Bioelectron. 91, 606–615 (2017). doi:10.1016/j.bios.2017.01.018.
(6) R.A. Lorenzo, A.M. Carro, C. Alvarez-Lorenzo, and A.Concheiro, Int. J. Mol. Sci. 12, 4327–4347 (2011). doi:10.3390/ijms12074327.
(7) S. Boulanouar,S. Mezzache, A. Combès, and V. Pichon, Talanta 176, 465–478 (2018). doi:10.1016/j.talanta.2017.08.067.
(8) M. Lasáková and P.J. Jandera, Sep. Sci. 32, 799–812 (2009). doi:10.1002/jssc.200800506.
(9) S. Ansari and M. Karimi, Talanta 167, 470–485 (2017). doi:10.1016/j.talanta.2017.02.049.
(10) L. Chen, S. Xu, and J. Li, Chem. Soc. Rev. 40, 2922 (2011). doi:10.1039/c0cs00084a.
(11) G. Vasapollo, R. Del Sole, L. Mergola, M.R. Lazzoi, A. Scardino, S. Scorrano, and G. Mele, Int. J. Mol. Sci. 12, 5908–5945 (2011). doi:10.3390/ijms12095908.
(12) E. Turiel and A. Martín-Esteban, Anal. Chim. Acta 668, 87–99 (2010). doi:10.1016/j.aca.2010.04.019.
(13) J. Haginaka, J. Sep. Sci. 32, 1548–1565 (2009). doi:10.1002/jssc.200900085.
(14) J.C. Masini and F. Svec, Anal. Chim. Acta 964, 24–44 (2017). doi:10.1016/j.aca.2017.02.002.
(15) M.M. Moein and M. Abdel-Rehim, Bioanalysis 7, 2145–2153 (2015), doi:10.4155/bio.15.153.
(16) M.R. Gama and C.B. Bottoli,J. Chromatogr. B 1043, 107–121 (2017). doi:10.1016/j.jchromb.2016.09.045.
(17) D. Stevenson, H.F. EL-Sharif, and S.M. Reddy, Bioanalysis 8, 2255–2263 (2016). doi:10.4155/bio-2016-0209.
(18) L.M. Madikizela, N.T. Tavengwa, and L. Chimuka, J. Pharm. Biomed. Anal. 147, 624–633 (2018). doi:10.1016/j.jpba.2017.04.010.
(19) D. Bitas and V. Samanidou, Molecules 23, 316–348 (2018). doi:10.3390/molecules23020316.
(20) L. Figueiredo, G.L. Erny, L. Santos, and A. Alves, Talanta 146, 754–765 (2016). doi:10.1016/j.talanta.2015.06.027.
(21) J.C.C. Yu and E.P.C. Lai, Toxins 2, 1536–1553 (2010). doi:10.3390/toxins2061536.
(22) V. Pichon and A. Combès, Anal. Bioanal. Chem. 408, 6983–6999 (2016).doi:10.1007/s00216-016-9886-0.
(23) S. Vogliardi, M. Tucci, G. Stocchero, S.D. Ferrara, and D. Favretto, Anal. Chim. Acta 857, 1–27 (2015).doi:10.1016/j.aca.2014.06.053.
(24) L.X. Yi, R. Fang, and G.H. Chen, J. Chromatogr. Sci. 51, 608–618 (2013). doi:10.1093/chromsci/bmt024.
(25) V. Pichon and F. Chapuis-Hugon, Anal. Chim. Acta 622, 48–61 (2008).doi:10.1016/j.aca.2008.05.057.
Dimitrios Bitas and Victoria Samanidou are with the Laboratory of Analytical Chemistry, Department of Chemistry, at Aristotle University in Thessaloniki, Greece. Direct correspondence to: samanidu@chem.auth.gr

New TRC Facility Accelerates Innovation and Delivery
April 25th 2025We’ve expanded our capabilities with a state-of-the-art, 200,000 sq ft TRC facility in Toronto, completed in 2024 and staffed by over 100 PhD- and MSc-level scientists. This investment enables the development of more innovative compounds, a broader catalogue and custom offering, and streamlined operations for faster delivery. • Our extensive range of over 100,000 high-quality research chemicals—including APIs, metabolites, and impurities in both native and stable isotope-labelled forms—provides essential tools for uncovering molecular disease mechanisms and exploring new opportunities for therapeutic intervention.
New Guide: Characterising Impurity Standards – What Defines “Good Enough?”
April 25th 2025Impurity reference standards (IRSs) are essential for accurately identifying and quantifying impurities in pharmaceutical development and manufacturing. Yet, with limited regulatory guidance on how much characterisation is truly required for different applications, selecting the right standard can be challenging. To help, LGC has developed a new interactive multimedia guide, packed with expert insights to support your decision-making and give you greater confidence when choosing the right IRS for your specific needs.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)









