An Improved Method for Eliminating Ion Suppression and Other Phospholipid-Induced Interferences in Bioanalytical Samples
Special Issues
This article examines the various effects of phospholipids in liquid chromatography tandem mass spectrometry (LC–MS-MS) analysis and demonstrates a new phospholipid-removal approach.
Phospholipids are perhaps one of the most troublesome components in bioanalytical samples when performing liquid chromatography coupled to tandem mass spectrometry (LC–MS-MS) analysis. Shortened column life, ion suppression, and an increase in MS system maintenance are just a few of the downfalls that can occur if phospholipids are not sufficiently removed from bioanalytical samples before analysis. Rapid crude sample preparation methods such as protein precipitation are sometimes preferred over more focused sample cleanup techniques like solid-phase extraction (SPE); however, such crude sample preparation techniques primarily reduce protein content and only slightly reduce the phospholipids present in the sample. Method developers are faced with a dilemma: Choose a sample preparation method that is quick but prone to method interferences because of the presence of phospholipids, or develop an optimized SPE method to achieve a cleaner, phospholipid-free sample. This article examines the various effects that phospholipids play in LC–MS-MS analysis and demonstrates a new phospholipid-removal approach that is simple and rapid like protein precipitation, yet removes phospholipids similar to a SPE procedure, but without the required method development.
In the past, the analysis of drugs, metabolites, and toxins in biological fluid was performed using a developed sample preparation technique that included either solid-phase extraction (SPE) or liquid–liquid extraction (LLE) before the sample was analyzed by liquid chromatography–mass spectrometry (LC–MS). As the size and complexity of studies increased, many groups undertook efforts to streamline methods and increase throughput by reducing LC run times and moving away from complex sample preparation methods. Indeed, many groups have simplified sample preparation methods such that extraction methods have been completely abandoned and a simple protein-precipitation method is used before LC–MS analysis. In addition, LC–MS run times have been reduced so that the chromatography is little more than a desalting step. Unfortunately, such reductions in run time and sample preparation have not come without encountering issues with sample matrices.
Interference caused by phospholipids in biological fluid samples has been a long-standing topic of debate because numerous groups have repeatedly demonstrated that most ion suppression in LC–MS-MS analysis of protein precipitated and direct inject sample is because of the presence of phospholipids. It has also been observed that phospholipids can cause several other negative effects in LC–MS-MS analysis such as a decrease in high performance liquid chromatography (HPLC) or ultrahigh-pressure liquid chromatography (UHPLC) column lifetimes, a decrease in MS sensitivity, and a subsequent increase in MS maintenance because of build up of nonionized lipids on the MS ion source.
These many adverse effects of phospholipids on LC–MS-MS analysis have spurred an increase in targeted sample preparation to selectively remove phospholipids from bioanalytical samples. In the past, it was difficult to selectively remove phospholipids because they exist in several compound classes with unique chemical and retentive properties. Of particular concern and interest in rapid LC–MS analyses are the phosphatidyl cholines and lysophosphatidyl cholines. Both classes of phospholipids can be visualized by LC–MS-MS by monitoring the 184→184 mass transition. Using this approach, this study looks at the effects of phospholipids in plasma samples following the use of two different sample preparation techniques: a standard protein-precipitation method and a simultaneous protein-precipitation and phospholipid-removal method using a 96-well phospholipid-removal plate.
Experimental Conditions
All solvents and buffer reagents were purchased from EMD. Laboratory chemicals were obtained from Sigma Chemicals and plasma samples were purchased from Bioreclamation.
Plasma samples were prepared using the two sample preparation techniques described in Table I. A standard protein-precipitation method was compared to a slightly modified procedure using a 96-well phospholipid-removal plate, Phree (Phenomenex). Each sample preparation technique called for up to five simple steps and required no sample specific method development outside the listed protocol. In an effort to maintain equivalence between methods, plasma samples from the same lot were used for each cleanup method, which ensured that the starting level of phospholipids was equivalent in each prepared sample.

Table I: Sample preparation protocols
After cleanup, samples were injected onto an Agilent 1200 HPLC system, which included an autosampler, solvent degasser, and column oven. The equipment was operated using Chemstation software (Agilent). The UHPLC column used was a 50 mm × 2.1 mm, 2.6-μm Kinetex C18 core–shell column (Phenomenex) that was coupled to an API 3000 mass spectrometer (AB Sciex). Phospholipids in each sample were then monitored using m/z 184–184. Mobile-phase A was 0.1% formic acid in water and mobile-phase B was 0.1% formic acid in acetonitrile. A gradient consisting of 5–95% B in 5 min or 15 min (Figures 1 and 2, respectively) was used, and the flow rate was 0.35 mL/min.
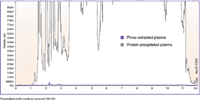
Figure 1: Comparison between protein-precipitated plasma and phospholipid-removal plate (Phree) extracted plasma samples. The MS-MS MRM transition for phospholipids is monitored across a gradient on a Kinetex C18 column. Note that phospholipid-removal plate samples exhibit a dramatic reduction in MS signal because of the selective removal of phospholipids.
Results
An overlay between the phospholipid-focused LC–MS analysis of protein-precipitated plasma sample and the same sample using a phospholipid-removal plate is shown in Figure 1. Contrary to some that report phospholipids only being at the beginning and end of a gradient, when a focused analysis is performed one can see phospholipids eluted across the entire UHPLC gradient in the protein-precipitation sample.
Even though phospholipids can be seen across the whole gradient when analyzing the low-level MS signal on the C18 column, their presence does not necessarily impact quantitation of all compounds in plasma samples. To better study the influence of ion suppression on the quantitation of an analyte, amoxapine was infused post-column to establish an ion suppression and enhancement profile in each of the prepared samples. Two mass transitions were then analyzed, m/z 184–184, which showed the total phospholipid profile, and m/z 314–271, which monitored for the amoxapine that was introduced via post-column infusion (Figure 2). Dramatic changes in the MS baseline (up or down) are indicative of suppression or enhancement effects of phospholipids in a plasma sample.
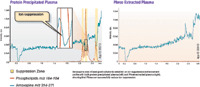
Figure 2: Ion suppression effect across an LC gradient for (a) protein-precipitated plasma and (b) plasma treated using a phospholipid-removal plate. Amoxapine post-column infusion maintains a 314â271 MS-MS signal across the gradient. Any rapid deviations (peaks or valleys) from baseline are indicative of ion suppression because of phospholipids. Note that with the protein-precipitated sample, a major depression in amoxapine signal at 2.4 min retention time corresponds to the phospholipids peak at the same time. The same sample run on the phospholipid-removal plate shows minimal signal suppression across the entire gradient.
In addition to the deleterious influences on analyte quantitation, a common finding from groups using protein precipitation has been that HPLC column lifetimes have been greatly reduced compared to columns used with other sample preparation techniques. A lifetime study was performed to determine the effects each sample preparation technique (protein precipitation vs. phospholipid-removal plate) had when multiple samples were repeatedly injected onto the LC–MS system. Repetitive 20-μL injections of diclofenac in protein-precipitated plasma versus diclofenac in phospholipid-removal plate extracted plasma (as prepared in Table I) were made onto the LC–MS system (Figure 3).
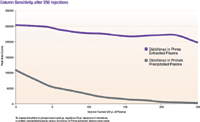
Figure 3: Column sensitivity after 250 injections.
Discussion
Our study revealed that the protein-precipitated plasma sample contained a significant amount of phospholipids (phosphatidyl cholines and lysophosphatidyl cholines) when monitoring for m/z 184–184. When monitoring the same mass transition, the phospholipid-removal plate extracted plasma samples showed a significant decrease in the presence of phospholipids to the point where they were virtually eliminated.
After confirming the presence and absence of phospholipids between protein-precipitation and phospholipid-plate extracted plasma, respectively, ion suppression in each prepared sample was studied. It was shown that a large region of suppression occurred in the protein-precipitated plasma when monitoring m/z 314–271 (amoxapine) (Figure 2). This region of suppression directly corresponded to a spike in phospholipid signal that was monitored using m/z 184–184. To confirm that phospholipids were the cause of the observed ion suppression, the same post-column infusion study was performed on the phospholipid-removal plate extracted plasma that we previously determined had been stripped of virtually all phospholipids. This study did not show a region of ion suppression and instead displayed a steady signal for the amoxapine infusion. It was therefore determined that the presence of phospholipids in the protein-precipitated sample caused a region of ion suppression in the amoxapine infused sample that did not occur in the sample that had been stripped of phospholipids.
Ion suppression is not the only negative effect that phospholipids can cause in LC–MS-MS analysis. Column lifetime and MS sensitivity are also at risk, so we studied the effect of phospholipids on each. It was shown that the phospholipid-removal plate extracted sample immediately yielded a signal that was 2.5 times stronger than the protein precipitated sample when the first injections were made. After repetitive injections, the signal from the protein-precipitated samples quickly decreased by more than 100,000 peak area counts to virtually zero after 250 injections. On the other hand, the signal from the phospholipid-removal plate extracted samples decreased only slightly, by 50,000 peak area counts after 250 injections (Figure 3).
Conclusion
Phospholipids are thought to play many negative roles in LC–MS analysis. Our study found that by using the correct sample preparation technique, phospholipids can be removed from samples before being injected onto an HPLC or UHPLC column. Protein precipitation, while a quick and easy method, was not able to remove phospholipids and resulted in several downfalls including ion suppression and an immediate decrease in sensitivity and column lifetime. Using a phospholipid-removal device, samples could be prepared using the same steps required in a traditional protein precipitation; however, the approach provided the added benefit of removing phospholipids. Extracted samples showed virtually no sign of phospholipids and immediately provided a 2.5 increase in sensitivity as well as an increased column lifetime compared to protein precipitated samples.
Stuart Kushon and Erica Pike are with Phenomenex in Torrance, California. Please direct correspondence to: EricaP@phenomenex.com.

Study Explores Thin-Film Extraction of Biogenic Amines via HPLC-MS/MS
March 27th 2025Scientists from Tabriz University and the University of Tabriz explored cellulose acetate-UiO-66-COOH as an affordable coating sorbent for thin film extraction of biogenic amines from cheese and alcohol-free beverages using HPLC-MS/MS.
Quantifying Microplastics in Meconium Samples Using Pyrolysis–GC-MS
March 26th 2025Using pyrolysis-gas chromatography and mass spectrometry, scientists from Fudan University and the Putuo District Center for Disease Control and Prevention detected and quantified microplastics in newborn stool samples.










