Capitalizing on the Benefits of Ion Mobility for Metabolite Identification
The goal of metabolite identification groups is to de-risk compounds moving into development by ensuring they have favourable metabolic profiles before clinical trials are initiated. Liquid chromatography–mass spectrometry (LC–MS) is a well-established technology for this purpose, as a result of its ability to selectively and accurately distinguish drugs and their metabolites. Ion mobility spectrometry (IMS) can add another critical dimension of separation by improving spectral clarity and generating collision cross section (CCS) values to track metabolites across multiple analysis conditions. Modern software platforms have now evolved to solidify the value of IMS for the field of metabolite identification.
Photo Credit: Orawan/stock.adobe.com

The goal of metabolite identification groups is to de-risk compounds moving into development by ensuring they have favourable metabolic profiles before clinical trials are initiated. Liquid chromatography–mass spectrometry (LC–MS) is a well-established technology for this purpose, as a result of its ability to selectively and accurately distinguish drugs and their metabolites. Ion mobility spectrometry (IMS) can add another critical dimension of separation by improving spectral clarity and generating collision cross section (CCS) values to track metabolites across multiple analysis conditions. Modern software platforms have now evolved to solidify the value of IMS for the field of metabolite identification.
Metabolite identification plays a crucial role in drug development, leading to a constant need to evaluate new technologies that can improve data quality or increase efficiencies. The recent example of a multiple sclerosis drug submitted for FDA approval, and the agency’s subsequent refusal to consider the application, illustrates the risk of incomplete characterization of clinically relevant metabolites (1). Additional technologies, such as ion mobility spectrometry (IMS), continue to gain attention in efforts to avoid complications and answer critical questions earlier in the process.
The metabolite identification challenges that technologies such as IMS can address depend on the drug development stage. Discovery laboratories need to identify key major metabolites and metabolic profiles for potentially hundreds of compounds in a short time frame. Preclinical development groups will need to more thoroughly characterize fewer numbers of compounds over a longer period of time, with efficient processes in place to meet the higher standard for reporting (2). In addition, technical methods developed for traditional therapies, such as small molecules, might not necessarily translate to newer biotherapeutic development (3). However, the biggest challenge might be balancing the workflow with the quality of the acquired data.
LC–MS Technologies and Ion Mobility in Metabolite Identification
Liquid chromatography–mass spectrometry (LC–MS) helps to address these challenges by separating compounds in mixtures using two dimensions, namely retention time (RT) and mass-to-charge ratio (m/z). Careful and reproducible analytical measurement of both properties usually allows the DMPK scientist to determine what overall biotransformations have occurred and what the structures look like over many samples or studies.
A variety of LC–MS instrumentation can be used for metabolite identification (4), including tandem quadrupole, quadrupole time-of-flight (QTOF), and orbital trap. Correspondingly, different acquisition methods are also employed, such as dataâdependent and data-independent acquisition (DDA and DIA, respectively). DDA methods, such as MS2, typically involve generating an inclusion list of likely metabolites a priori so that as many metabolites as possible are identified and collected with useful fragmentation information. DDA reduces processing time and file size, while providing good, quality spectra for expected molecules. The potential downside is missing unexpected metabolites that are not part of the inclusion list, a critical risk in metabolite identification (5). DIA methods, such as full scan or MSE, solve this problem by generating information on all components for all product ions. This reduces the potential for missed metabolites, but can also be timeâconsuming because of the amount of data produced (6). MSE rapidly alternates between full scan low collision and high collision energies, allowing both precursor (drug and metabolite) and product ion high resolution accurate mass measurements to be obtained.
IMS has now evolved to afford an added dimension of separation to metabolite identification, based on an ion’s size and shape, along with an accurate mass-toâcharge ratio, as it passes through a gas such as nitrogen. This property can be used to generate a collision cross section (CCS) value, correlated to its drift time (DT), that describes the ion’s overall size (influenced by structure, charge, conformation, and interaction with the gas) (7). When combined with MSE, this acquisition method generates high definition MSE (HDMSE) data. The development of IMS is decades-old, but its availability in combination with commercialized LC–MS instruments has led to increased interest in its relevance for a range of applications, including metabolite identification (8,9). Ion mobility supported on commercial LC–MS platforms now includes traveling wave ion mobility (TWIMS), drift tube ion mobility (DTIMS), and trapped ion mobility (TIMS), all of which generate CCS values that can be stored in customized libraries. Differential ion mobility (DMS) and high field asymmetric waveform ion mobility (FAIMS) improve selectivity; however, they do not generate CCS values. Each of the ion mobility techniques generates quality data, but DIA ensures that no metabolites are missed (10). In addition, the data can be stored and reâinterrogated at a later time point or used to compare across experiments.
IMS improves data quality and adds another dimension of separation for the identification and structural elucidation of metabolites, as shown in Figure 1 (11). Spectral clarity is improved by using DT to align retention time data and filter out nonpertinent ions, which is especially important when looking for metabolite fragment ions in biological matrices of preclinical toxicity models (12). In addition, the precise CCS value, which is matrix and ion-concentration independent, can be used to track isomeric metabolites across chromatographic conditions and resolve coeluting compounds throughout the entire drug development life cycle (13). A single compound will spend years progressing through development, being tested in increasingly more complex biological systems, and resulting in more detailed data sets. This ability to more confidently detect, characterize, and track metabolites across time and geographies without worrying about varying chromatographic conditions greatly eases the burden of the biotransformation scientist.
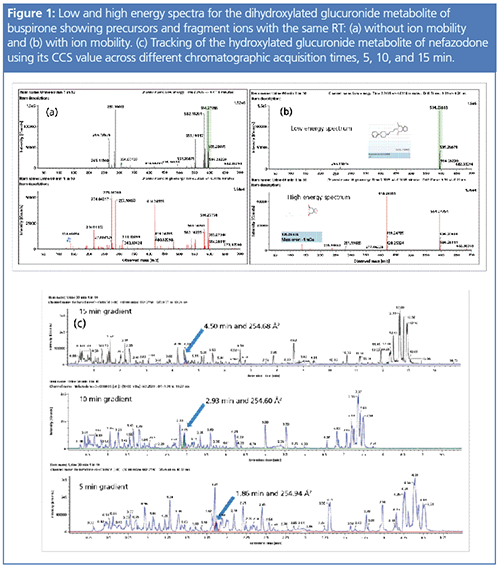
IMS adds value for the biotransformation scientist, no matter the drug development stage. In discovery, the key biotransformation pathways of a compound must be understood, and chemical groups that are liable or have potential for toxicity must be replaced. If this is not done accurately, it is very difficult to determine if a metabolite originated from or via a reactive intermediate pathway and this can lead to costly reanalysis. As the compound moves into preclinical development, the concern is with comparing metabolites formed across species in order to choose or validate a model toxicity species that can represent human metabolism. Better spectral clarity through IMS greatly improves the scientist’s ability to precisely identify and elucidate all metabolite structures throughout drug development. In addition, the ability to track or assign the metabolite with more confidence across different matrices and species using both mass and CCS as a drug moves towards definitive human studies is critical.
The Importance of Software for Ion Mobility Data Interpretation
LC–MS in combination with IMS generates high-quality data that directly impacts the confidence of later conclusions; however, software is needed to easily manage, analyze, and store these complex data sets efficiently. Fortunately, several solutions have been developed that incorporate IMS into software packages at each stage of development and across molecule types (14).
In discovery, the algorithms used to make metabolic predictions must be suited to high throughput, multicompound, batch processing. In order to de-risk a candidate compound in a lead optimization program, that information must be fed back into the process in an efficient manner for the next round of compound design. In development, throughput needs decrease, but compliance, audit trails, and fit-for-purpose reporting become increasingly important.
Molecule type is also a consideration when choosing the appropriate software platform. Compared to traditional small molecule development, metabolite identification software for peptides must address a larger number of potential cleavages, have more predictable rules than for small molecules, and must be able to handle both linear and cyclic forms (15). Software programs that characterize clearance and metabolic fate of these biotherapeutics are quickly being developed with improved functionality.
Many software programs for metabolite identification are exclusively for use with specific vendor instrumentation, integrating their technology or acquisition method into the overall workflow (16). Third party software, on the other hand, is intentionally agile and vendor agnostic so that it can be used as a common platform in laboratories that might have a variety of equipment vendors (17). In these situations, databasing of outcomes from multiple compounds across multiple conditions using a cloud-based system is valuable because it allows data to be shared, mined, and analyzed across experiments and laboratory sites.
The ultimate result of combining ion mobility data with the appropriate software is a more efficient, cost-effective workflow in the DMPK lab that can increase productivity and decrease the time it takes to handle, process, and analyze information, leaving the skilled biotransformation scientist more time and flexibility to add value on more difficult projects. Software programs geared to discovery can automatically calculate formal metabolic clearance outcomes, model the most likely site of oxidation, identify structural determinants of enzyme interactions, and suggest isosteric replacements for metabolically labile molecules. In addition, drug metabolites in biological samples can be identified across time points and structures proposed (Figure 2) (18), with these results databased and visualized to enable data sharing, mining, and new chemistry design, all within the needed time frame.
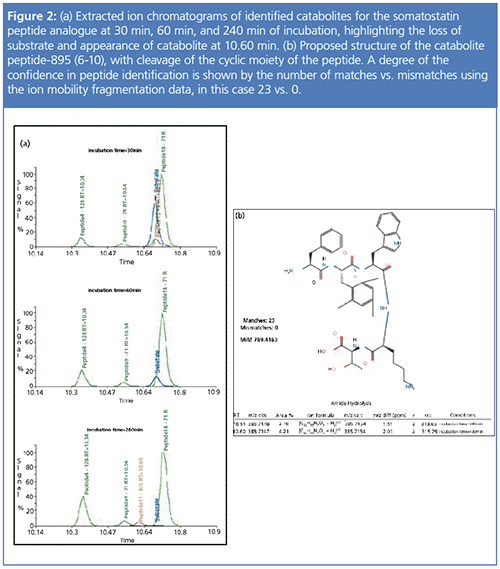
Conclusion
New technologies for metabolite identification workflows, such as ion mobility, are needed to generate the best data, within the needed time frame, at the appropriate phase of the drug discovery and development process. IMS delivers this capability by adding another separation dimension that increases confidence in identification and structural elucidation, distinguishes coeluting metabolites, and tracks metabolites across different matrices and chromatographic conditions using CCS values. When combined with the effective software, ion mobility can improve efficiencies and workflows to deliver real value to the metabolite identification laboratory.
References
- https://www.biopharmadive.com/news/celgene-ozanimod-metabolite-safety-test-approval/522268/
- G. Webber, Drug Discovery World Spring 2017.
- L. Di, The AAPS Journal17(1), 134–143 (2015). doi:10.1208/s12248-014-9687-3.
- N. Iwamoto and T. Shimada, Pharmacology and Therapeutics185, 147–154 (2018).
- R.L. Last, A.D. Jones, and Y. Shachar-Hill, Nat. Rev. Mol. Cell. Biol.8, 167–174 (2007).
- K.P. Bateman et al., Rapid Commun. Mass Spectrom.21, 1485–1496 (2007).
- C.R. Goodwin, L.S. Fenn, D.K. Derewacz, B.O. Bachmann, and J.A. McLean, J. Nat. Prod.75(1), 48–53 (2012). doi: 10.1021/np200457r
- C. Lapthorn, F. Pullen, and B.Z. Chowdhry, Mass Spectrom. Rev. 32(1), 43–71 (2013). doi: 10.1002/mas.21349
- T. Katsila, A.P. Siskos, and C. Tamvakopoulos, Mass Spectrom. Rev.31(1), 110–133 (2012). doi: 10.1002/mas.20340
- U. Distler, J. Kuharev, P. Navarro, Y. Levin, H. Schild, and S. Tenzer, Nature Methods11(2), 167–175 (2014). doi:10.1038/nmeth.2767
- J. Kirk, R. Mortishire-Smith, and M. Wrona, Waters Application Note 7200061 21EN (2017).
- S. Blech and R. Laux, Int. J. Ion Mobil. Spec.16, 5 (2013). https:/doi.org/10.1007/s12127-012-0113-1
- C. Holdsworth, R. Clayton, H. Robinson, C. Lord-Mears, and J. Kendrick, “Utilization of Ion Mobility Enabled Collisional Cross Section Measurements for the Comparison of Metabolites across Differing Chromatographic Methods,” poster presented at the Joint DMDG/GMP Open Meeting, Paris, France, 2016.
- A. Paiva and W. Shou, Bioanalysis8(16), 1723–1733 (2016).
- T. Radchenko, A. Brink, Y. Siegrist, C. Kochansky, A. Bateman, F. Fontaine, et al., PLoS ONE 12(11), e0186461 (2017).
- B. Bonn, C. Leandersson, F. Fontaine, and I. Zamora, Rapid Commun. Mass Spectrom.24(21), 3127–3138 (2010). doi: 10.1002/rcm.4753
- A. Brink, Drug Discov. Today Technol.10(1), e207–217 (2013). doi: 10.1016/j.ddtec.2012.12.001
- J. Kirk, I. Zamora, A. Riera, T. Radchenko, A. Escola, Y. Alelyunas, R. Mortishire-Smith, and M. Wrona, “Characterising the Catabolism of Peptides Using Ion Mobility Enabled High Resolution Mass Spectrometry with Mass-MetaSite Integration for Data Processing,” poster presented at the 23rd International Mass Spectrometry Conference, Florence, Italy, 2018.
Nathan Anderson is a business development manager at Waters Corporation, focusing on discovery and DMPK in the pharmaceutical market. He holds an M.B.A. from Boston University’s Questrom School of Business (Boston, MA, USA) and an M.S. in biology from American University (Washington, DC, USA).
E-mail:Nathan_Anderson@waters.comWebsite:dmpk.waters.com

HPLC 2025 Preview: Fundamentally Speaking (Part 1)
May 13th 2025Michael Lämmerhofer from the Institute of Pharmaceutical Sciences, University of Tübingen, Germany, spoke to JFK Huber Lecture Award winner of 2024 Torgny Fornstedt, professor in analytical chemistry and leader of the Fundamental Separation Science Group, Karlstad University, Sweden, about his pioneering work in high performance liquid chromatography (HPLC) with a focus on fundamentals and industrial applications.
Reversed-Phases for LC Deliberately Doped with Positive Charge: Tips and Tricks for Effective Use
May 13th 2025In this month's edition of LC Troubleshooting, Dwight Stoll and his fellow researchers discuss both the benefits (improved peak shape/loading) and challenges (excessive interaction) associated with charge-doped reversed-phase (RP) columns for both analytical and preparative separations.
Determining Ways to Protect Honeybee Colonies with GC–MS
May 13th 2025A study conducted by the Agriculture Research Centre of Giza, Egypt, and Jilin Agricultural University in China, evaluated the efficacy of stinging nettle extract, nettle smoke, and formic acid in the controlling of Varroa mites, a major threat to honeybee colonies, with a focus on mite infestation reduction, honeybee mortality, and biochemical responses. Gas chromatography–mass spectrometry (GC–MS) was used to identify key bioactive compounds in the stinging nettle extract.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)












