An Age of Discovery
LCGC spoke to Martin Giera of Leiden University Medical Centre, Leiden, The Netherlands, about developments in biomarker discovery.
LCGC spoke to Martin Giera of Leiden University Medical Centre, Leiden, The Netherlands, about developments in biomarker discovery.
Q: Your group is actively involved in biomarker discovery. When did you become involved in this field and how has it evolved since you first started?
A: I got involved in biomarker discovery around two years ago. During this time I would say that speed is becoming more and more important, especially because studies are becoming larger and larger, which naturally involves increased sample numbers.
Q: What are the main chromatographic techniques that you use and what are the biggest challenges you face in biomarker research generally?
A: Our group uses different separation technologies, ranging from well‑established, robust methods such as gas chromatography (GC) to a relatively experimental set-up based on capillary electrophoresis. However, HPLC — either in a capillary set-up or in combination with 2-mm core-shell columns — remains our main chromatographic technique. Core shell columns have proved to be very useful because they provide close to UHPLC performance, but we can also use our standard LC equipment.
Q: Your group has focused on derivatization strategies in biomarker discovery. Why is derivatization still important in this field and is this approach time-consuming?
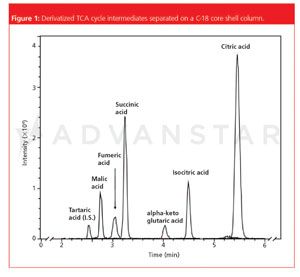
A: Most biomarker studies today are performed with LC–MS platforms operated with a RP-18 or HILIC column. Of course, this will restrict the analytes that can possibly be detected to what is retained on the incorporated columns. Many analytes also show restricted ionization capabilities that makes their detection even more difficult. Hence we asked the question if it would be better to derivatize certain analyte subclasses for better chromatographic separation and enhanced detection. Even though this might lead to more bias in the analysis it might allow the detection of metabolites that would otherwise be overlooked. One example where we enhanced the detection limits by a factor of almost ten with a derivatization strategy was for the analysis of small carboxylic acids, including the intermediates of the tri-carboxylic acid cycle. In addition to this, alternate isotope-coded derivatization (AIDA) and group specific internal standard technology (GSIST) approaches can provide convenient quantification in combination with derivatization strategies.
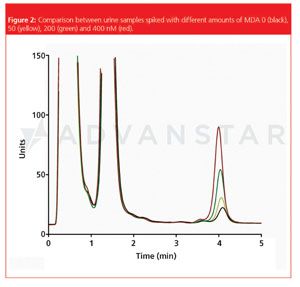
Another important field of derivatization is the selective derivatization of established biomarkers. The main goal here is the highly sensitive and fast determination of such substances. In this field we recently conducted a study aimed at the determination of malondialdehyde: an important reactive carbonyl, a biomarker of oxidative stress and a side-product of thromboxane synthesis. With the developed labeling strategy we were able to detect malondialdehyde down to concentrations of around 2 nM in urine samples while the chromatographic run times were shorter than 5 min. Crucial for the separation of highly fluorescent urinary components and our derivatized analyte was the use of a cyano-column which we used in the reversed-phase mode.
Q: You are involved in some interesting work relating to human infectious diseases such as urinary tract infections and tuberculosis. How does this work benefit patients and what was the biggest challenge you had to overcome in terms of chromatography?
A: Our project on Urinary Tract Infections is part of a collaboration between our group and the department of Infectious Diseases at the LUMC (headed by Professor Jaap van Dissel). We are aiming to find markers for the identification of the causative bacteria. A current “golden standard” of pathogen identification is bacterial culture, which takes 24 h to 48 h. If we could develop a method that gives us an answer within, for example an hour, it could lead to a much more tailored treatment strategy and, most importantly, reduced risk for the development of bacterial resistance to the applied antibiotics.
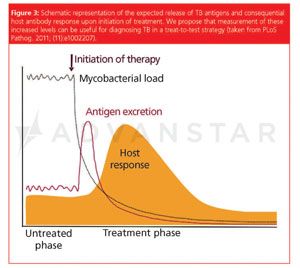
The project about tuberculosis biomarkers is a result of our collaboration with the KIT (Royal Dutch Tropical Institute, particularly Alice den Hertog and Richard Anthony). The goal of this study is to evaluate a recently proposed “treat‑to‑test strategy” where the treatment and consecutive response are used as the diagnostic tools. Well, it remains to be seen whether the “treat‑to‑test strategy” will work or not; our role in this project, however, is to identify a time window after the treatment when the pathogen‑related substances (metabolites) appear and provide as detailed as possible characterization of these substances.
In terms of chromatography: it is frequently about making sacrifices. If you are facing several hundred samples your primary goal is usually speed. This is why we tend to use core-shell particle columns. With respect to the column chemistry one has to decide whether to use HILIC or RP: both chemistries have their benefits and drawbacks. However, the well-established robustness of RP separations usually makes us use this column chemistry. For the future it will be interesting to see how C30 columns or mixed-mode phases might influence the field.
Q: What other projects are you excited about at the moment?
A: A very recent study which we performed in co-operation with Professor Charles N. Serhan from Harvard Medical School and the department of rheumatology at the LUMC was aimed at the analysis of lipid mediators in arthritis patients. In addition to the analysis of the lipid mediator class we also performed a general lipid profiling of the patient material. At present we are very excited about the results and are now planning to investigate the biological roles of the lipid mediator class in different biological assays further.
Q: How do you see this work evolving in the future?
A: I am pretty sure that sample numbers will keep getting larger, which will demand faster separations and extremely robust analysis platforms. For the biomarkers themselves I believe that their thorough validation is key. Additionally, it will also be crucial to develop, as far as possible, simple antibody- or derivatization-based tests that can easily be applied in the clinic by the doctors or nurses.
Q: Anything else you would like to add?
A: I’d like to thank all people involved in my past and present work, especially Charles N. Serhan from HMS, Andre M. Deelder, Oleg A. Mayboroda, Andreea Ioan-Facsinay and Rene Toes from the LUMC and my former colleagues at the VU University Amsterdam, Wilfried MA Niessen and Henk Lingeman.

Martin Giera studied pharmacy at the universities of Heidelberg and Munich, Germany. After a 7 month stay in the analytical department of Boehringer Ingelheim (Biberach Riss, Germany) he joined the group of Professor Bracher at the LMU Munich for his doctoral studies in pharmaceutical chemistry. He received his PhD in late 2007. In 2008 he joined the group of Professor Hubertus Irth at the VU University Amsterdam, The Netherlands, as a postdoc. In 2009 he was appointed assistant professor in the same group. Since May 2011 he is mainly responsible for the lipidomics activities in the biomolecular mass spectrometry unit of Professor Andre M. Deelder/Associate Professor Oleg A. Mayboroda at the Leiden University Medical Centre (LUMC).

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)




