Advances in Spatial Biology using TIMS-TOF MS
Spatial biology investigates cellular arrangements within tissues, providing insights into biological function and disease. The emergence of trapped ion mobility spectrometry-time of flight (TIMS-TOF) mass spectrometry (MS) has advanced this field by enabling the analysis of individual cells and subcellular compartments with unprecedented sensitivity. Unlike traditional MS, TIMS-TOF MS can extract detailed molecular information from tissues and organelles. This technology offers sub single-cell level sensitivity, crucial for examining small biological specimens such as immune cells or organelles. TIMS-TOF MS facilitates the integration of multiple omics technologies, creating a comprehensive toolbox that combines spatial and molecular data. While traditional spatial biology methods rely on RNA and DNA, TIMS-TOF directly assesses biological function, bypassing genomic intermediates.
Researchers can now apply spatial omics data from matrix assisted laser desorption ionization (MALDI) imaging on tissue sections with LC hyphenated to trapped ion mobility time-of-flight mass spectrometry (LC–TIMS-TOF MS) experiments. These approaches integrate mass spectrometry imaging (MSI) with LC–MS in two main workflows: one where MSI guides LC–MS by targeting specific areas of interest within tissue sections for detailed analysis, and another where findings from LC–MS are used to inform MSI for the validation of a panel of biomarkers, facilitating rapid and multiplexed screening.
Innovative instruments and workflows have driven these developments, including improved spatial resolution, the integration of photocleavable mass tags in MALDI imaging, and increased speed and sensitivity in LC–MS analysis enhancing overall performance (1).
Many research groups suggest that this approach might eventually make the step from a research application into clinical practice—as a standardized workflow that proves the utility of translating research workflows into clinical application. This article illustrates the potential of these developments by summarizing recent published work from a series of expert groups.
Beyond Genomic Sequencing: Multiomics
In recent years researchers have moved beyond large-scale genomic sequencing to embrace a multiomic/systems-level approach combining the genome with, for example, ribonucleic acid (RNA) transcription (transcriptomics), proteins/peptides (proteomics), lipids (lipidomics), and metabolites (metabolomics).
This approach allows gene information to be considered together with complementary datasets to develop a more systemic understanding of diseases or other phenotypes of interest. MS has emerged as a cornerstone of omics analysis, continually expanding its potential through advances in instrumentation, sample preparation, software-guided workflows, and data processing capabilities. More recently, TIMS-TOF-MS has been adopted, adding an orthogonal separation dimension that enhances the ability to resolve isobaric species and improves confidence in molecular identification during the separation, identification, and quantitation of biological molecules. These developments have significantly deepened our knowledge of biological systems and disease etiology than genomics alone.
For research in many fields, scientists are now asking questions that extend beyond traditional approaches. They seek data to help understand the role of a particular molecule, or molecular change, in situ, in its native tissue environment. Despite the power of MS, the technique does not provide information about the location and physical distribution of the molecules under analysis, preventing researchers from directly overlaying the MS data with the tissues or cells of interest.
Enter MSI. Much has been written about the power of MALDI guided imaging combined with MS since it was first described back in 1997 (2). Commercial systems that enabled the import and co-registration of other imaging modalities, such as fluorescence or histopathology, became available in 2005 (3) and the technique has been described as painting molecular pictures (4).
It has served as the necessary spatial omics bridge between molecular analysis and imaging (Figure 1). Advancements in instrumentation and methodology have since allowed researchers to routinely investigate the precise locations of molecules of interest within tissue sections (Figure 2) – and even within individual cells.
FIGURE 1: Moving from genomics to spatial omics.
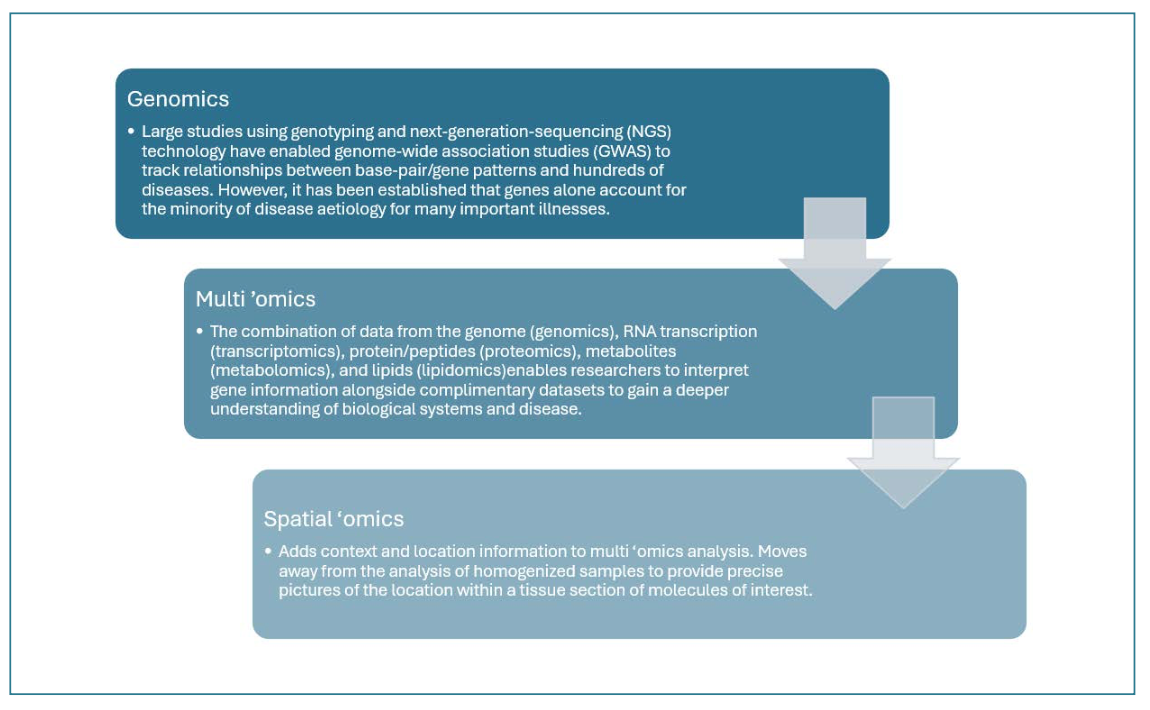
FIGURE 2: Four proteins distributed in rat brain.
Automated Laser Microdissection for Ultra-High-Resolution LC/MS Proteomics Reveals Molecular Pathways that Change as a Cancer Progresses
A recent study (5) aimed to combine sub-micron-resolution microscopy, image analysis for single-cell phenotyping based on artificial intelligence (AI), automated laser microdissection (LMD/LCM) and analysis with an ultra-sensitive proteomics workflow using LC–MS. The autosampler was configured for sample pick-up from 384-well plates, enabling high-throughput analysis of low volume samples.
The research used software to coordinate scanning and LMD microscopes. Cellular or subcellular objects of interest were selected by AI before being subjected to automated LMD and proteomic profiling.
They report that accurate definition of single-cell boundaries and cell classes, the transfer of the automatically defined features into proteomic samples ready for analysis, and robust, automated sample preparation workflows were key to achieving high sample throughput without compromising sensitivity. The precise definition of single-cell boundaries is particularly important in proteomics to reduce sample heterogeneity and enhance data quality.
Multiplexed Imaging for Ultra-High-Resolution LC/MS to Understand the Role of Immune Cell Infiltration in Cardiac Arrest
Spatial omics of acute myocardial infarction (MI), more commonly known as a heart attack, was studied in a multiomics workflow using TIMS-TOF technology to reveal a novel mode of immune cell infiltration (6).
Although MI is a leading cause of death worldwide, the precise changes in tissue architecture following an attack are poorly understood. This study utilized a combination of imaging-based transcriptomics (spatial transcriptomics, often referred to as molecular cartography) and antibody-based highly multiplexed imaging (sequential immunofluorescence) to evaluate cell-type compositions and changes at subcellular resolution.
The analysis identified a novel mode of leukocyte accumulation in the heart. The underlying mechanisms driving this previously unknown infiltration route were analyzed using deep visual proteomics (DVP), an unbiased spatial proteomic analysis. DVP enabled the identification of specific proteomic markers and pathways that were crucial to understanding this novel infiltration.
The approach led to the development of the first spatial map of acute murine MI with subcellular resolution. Furthermore, DVP was used to identify von Willebrand Factor (vWF) as a mediator of inflammation 24 hours post-MI—a previously unknown route of immune infiltration.
During an acute infarct, cells in the heart release stress signals that lead to the invasion of the infarct zone by immune cells. Modulation of these immune cells has been suggested as potential treatment targets to improve healing and outcome after MI, and a better understanding of the immune cell infiltration routes, detailed tissue microenvironment and cellular interactions in the heart hold the promise to deliver new treatment strategies.
MALDI HiPLEX Technology for Multiomic Profiling in a Single Experiment on a Single Slide
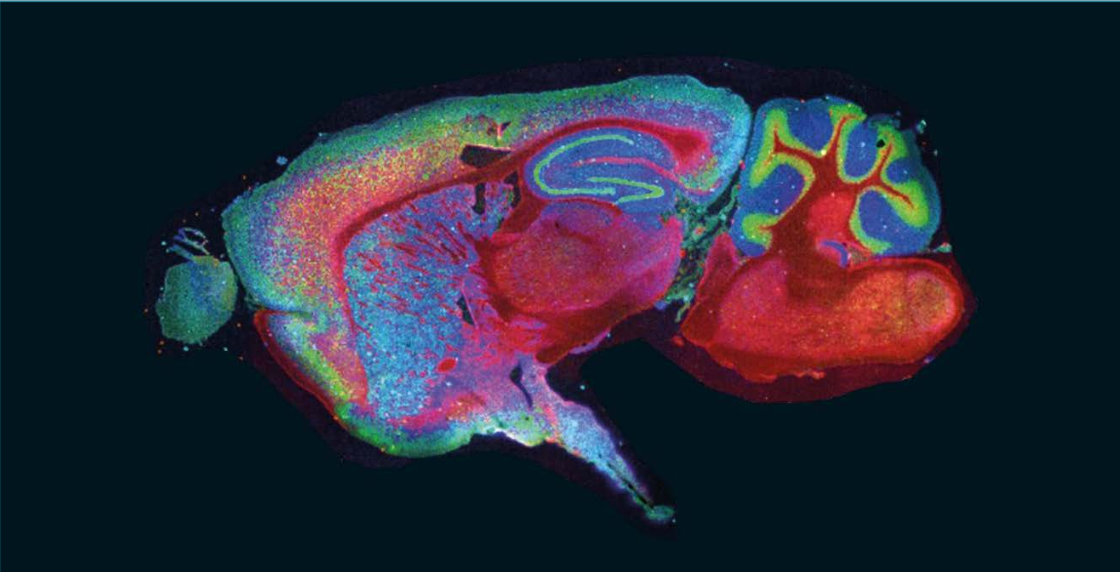
For many applications, it is important to note that MS, including TIMS-TOF MS and MALDI imaging, is a label-free technique; however, by using a range of available tags and labels, sensitivity and specificity can be significantly enhanced. Additionally, integrating photocleavable mass tags into a straightforward workflow with automated setup routines unlocks the potential for multiomic multiplex analysis on the same tissue section (Figure 3). These tags operate by releasing distinct mass tags upon irradiation, enabling the simultaneous detection of multiple biomarkers.
FIGURE 3: Multiomic imaging using photocleavable mass tags.
Multiomic analysis facilitates the determination of the localization and potential colocalization of a number of biomarkers simultaneously, which is important to drive research in oncology, for example, where it is critical in mapping the location of the hundreds of possible proteins involved in cell regulation and dysregulation in a highly heterogeneous tissue.
The standard method of multiplex immunohistochemistry (IHC), however, using standard fluorescence microscopy, is limited to between three and five different biomarkers and often require laborious cycling strategies. A new application, based on well proven photocleavable mass-tags that enable antibody labelling, facilitates highly multiplexed IHC based on MALDI mass spectrometric imaging, far exceeding the multiplexity of both fluorescence- and previous cleavable mass-tag-based methods (7).
This enables a high level of multiplexing without the limitations of optical methods. By generating a full mass spectrum at each pixel, the simultaneous imaging of any given mass species within the spectra is achieved (8). It is this combination of MALDI-MSI with IHC (known as MALDI-IHC) that overcomes the existing limitations (9), and allows for highly multiplexed imaging of targeted biomolecules in biospecimens.
MALDI-IHC combines the spatial resolution of MALDI imaging with the specificity of immunohistochemistry (IHC), enabling the visualization of spatially targeted protein expression across a large field of view within tissue sections. It supports multimodal workflows to overlay spatial proteomics data with distributions of lipids, endogenous peptides, or enzymatically released glycans or peptides, and to match those profiles with tissue histomorphology.
The technology can simultaneously image potentially hundreds of targeted biomarkers in FFPE and FF thin-tissue sections to provide more powerful methods for researchers to explore the spatial distribution of biomolecules in tissues at the cellular level in tissue pathology, tissue diagnostics, therapeutics and precision medicine.
MALDI Imaging – LCM – Proteomics Elucidate Molecular Profiles in Native Tissue Samples
The development of many analytical technologies has followed the same path: Researchers push the boundaries of an instrument or workflow because they demand more and more information from their sample—and companies respond with systems that offer more performance, simplified daily use with software-guided instrument set up, for example, and advanced data handling capabilities.
As noted above, to answer current questions researchers are moving past the analysis of a homogenized tissue sample to realize the value of analysis with spatial context.
Recent work by Professor Heeren at Maastricht University provides a good example of this groundbreaking approach. His group is studying molecular profiles at 5–10 µm in an effort to understand how spatially structured communities of individual cells act, and interact, in the context of their networked environment (10).
To achieve this, MALDI imaging guided spatial omics identifies regions of specific cell phenotype of interest on a slide, which can then be collected using LCM for proteomics study. In Maastricht, both experiments are conducted using the latest innovation in instrument design—a dual-source system, that has both MALDI and electrospray ionization (ESI) sources built in and promises state-of-the-art MSI and liquid-based proteomic capabilities on the same MS instrument.
This study clearly demonstrates that such an instrument can offer both fast lipid-based MSI at high mass and high lateral resolution, and sensitive LC–MS on local protein extracts from the same tissue section (11).
Conclusions
Analysts are constantly looking to boost performance of every element of the multiomics workflow with, for example, new multiplexed detection technology for proteomics, enabling analysis of an increased number of proteins to map cellular processes without increasing measurement time for a specified region of interest. Importantly, as applications become more sophisticated, the setup and operation of the instruments, as well as the ability to process and handle the large datasets generated, have significantly advanced.
A crucial driver for innovation is the potential to move from discovery and research to translate multiomics findings into a clinical context. While the vision of routine clinical application is likely still many years away, researchers are starting to explore proofs of concept for this next generation approach, paving the way for enhanced disease understanding and, in the longer term, improved diagnostics and targeted personalized treatment strategies.
The latest high-performance MALDI-TOF technology for MS-based tissue imaging is now available in an advanced benchtop system that offers rapid workflows for biopharmaceutical and clinical research with characterization at high hydrophobicity, heterogeneity or high speed. Moreover, integrating MALDI-TOF technology into a spatial biology workflow allows for pinpointing protein location and activity, building a greater understanding of both healthy and diseased tissue environments. By adopting a comprehensive approach from sample preparation to post-processing analysis, research in neuroscience, infectious disease and oncology can be transformed, and, in the longer term, drive new insights that will help to understand diseases in a more comprehensive way.
References
(1) Lemaire R.; Stauber J.; Wisztorski M.; Van Camp C.; Desmons A.; Deschamps M.; Proess G.; Rudlof I.; Woods A. S.; Day R.; Salzet M.; Fournier I. Tag-mass: specific molecular imaging of transcriptome and proteome by mass spectrometry based on photocleavable tag. J. Proteome Res. 2007, 6, 2057–2067. DOI: 10.1021/pr0700044.
(2) Caprioli, R. M.; Farmer, T. B.; Gile, J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem. 1997, 69 (23), 4751–4760. DOI: https://doi.org/10.1021/ac970888i.
(3) Lim M. J.; Liu Z.; Braunschweiger K. I.; Awad A.; Rothschild K. J. Correlated matrix-assisted laser desorption/ionization mass spectrometry and fluorescent imaging of photocleavable peptide-coded random bead-arrays. Rapid Commun. Mass Spectrom. 2014, 28, 49–62. DOI: 10.1002/rcm.6754.
(4) Schwamborn, K.; Caprioli, R. M. MALDI Imaging Mass Spectrometry-Painting Molecular Pictures. Mol. Oncol. 2010, 4 (6), 529–538. DOI: https://doi.org/10.1016/j.molonc.2010.09.002.
(5) Mund, A.; Coscia, F.; Kriston, A.; Hollandi, R.; Kovács, F.; Brunner, A.-D.; Migh, E.; Schweizer, L.; Santos, A.; Bzorek, M.; Naimy, S.; Rahbek-Gjerdrum, L. M.; Dyring-Andersen, B.; Bulkescher, J.; Lukas, C.; Eckert, M. A.; Lengyel, E.; Gnann, C.; Lundberg, E.; Horvath, P.; Mann, M. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nature Biotechnol. 2022, 1–10.
(6) Wünnemann, F, Sicklinger, F, Bestak, K, Nimo, J, Thiemann, T, Amrute, J, Nordbeck, M, Hartmann, N, Ibarra-Arellano, M. A, Tanevski, J, Heine, C, Frey, N, Lavine, K. J, Coscia, F, Saez-Rodriguez, J, Leuschner, F, Schapiro, D. Spatial omics of acute myocardial infarction reveals a novel mode of immune cell infiltration, bioRxiv, ASAP. DOI: 10.1101/2024.05.20.594955.
(7) Yagnik, G, Liu, Z, Rothschild, K.J, Lim, M. J. Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues. J. Am. Soc. Mass Spectrom. 2021, 32 (4), 977–988. https://pubs.acs.org/doi/10.1021/jasms.0c00473“https://pubs.acs.org/doi/10.1021/jasms.0c00473#
(8) Arentz, G.; Mittal, P.; Zhang, C.; Ho, Y. Y.; Briggs, M.; Winderbaum, L.; Hoffmann, M. K.; Hoffmann, P. Applications of Mass Spectrometry Imaging to Cancer. Adv. Cancer Res. 2017, 134, 27– 66, DOI: 10.1016/bs.acr.2016.11.002.
(9) Dewez F.; Oejten J; Henkel C; Hebeler R; Neuweger H; De Pauw E.; Heeren R.M.A; Balluff B. MS Imaging-Guided Microproteomics for Spatial Omics on a Single Instrument. Proteomics 2020, DOI: 10.1002/pmic.201900369.
(10) Claes, B. S. R., Krestensen, K. K., Yagnik, G., Grgic, A., Kuik, C., Lim, M. J., Rothschild, K. J., Vandenbosch, M., & Heeren, R. M. A. MALDI-IHC-Guided In-Depth Spatial Proteomics: Targeted and Untargeted MSI Combined. Anal. Chem. 2023, 95 (4), 2329–2338. DOI: 10.1021/acs.analchem.2c04220.
(11) Dewez, F.; Oejten, J.; Henkel, C.; Hebeler, R.; Neuweger, H.; DePauw, E.; Heeren, R. M. A.; Balluff, B. MS Imaging-Guided Microproteomics for Spatial Omics on a Single Instrument. Proteomics 2020, 20, 1900369. DOI: 10.1002/pmic.201900369.
About the Authors
Manuel Bauer is the VP of Product Management of Consumables & Automation at Bruker Daltonics.

Pascal Steffen-Lockhauserbäumer is a Product Manager timsTOF at Bruker Daltonics.

Jan H. Kobarg is a Product Manager for MALDI-TOF at Bruker Daltonics.


New Method Explored for the Detection of CECs in Crops Irrigated with Contaminated Water
April 30th 2025This new study presents a validated QuEChERS–LC-MS/MS method for detecting eight persistent, mobile, and toxic substances in escarole, tomatoes, and tomato leaves irrigated with contaminated water.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)








