Machine Learning and MALDI-TOF-MS for Predicting Antibiotic Resistance
Universidad Católica del Maul scientists reviewed advancements made in using machine learning and MALDI-TOF-MS for predicting antimicrobial resistance in different materials.
Scientists from Universidad Católica del Maule in Talca, Chile recently reviewed advancements made in using machine learning (ML) and matrix-assisted laser desorption/ionization time-of-flight mass spectra for predicting antimicrobial resistance (AMR) in different materials. Their findings were published in the Journal of Chromatography A (1).
Antibiotic resistance has become a serious global health issue in recent years. Described as when bacteria change so that antibiotic medicines cannot kill or stop their growth, this phenomenon has become associated with higher morbidity and mortality rates (2). In the United States, more than 23,000 deaths annually are associated with antibiotic resistance, and the World Health Organization (WHO) has predicted that by 2050, antibiotic resistance will cause more than 10 million human deaths. This public health crisis has mainly been attributed to excessive and indiscriminate antibiotic prescription from medical professionals, increased antibiotic use in food production (livestock, poultry, fishery, and agriculture), and environmental factors.
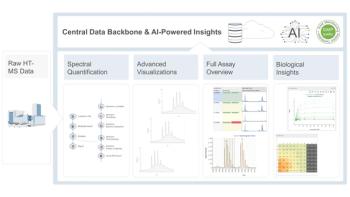
This issue partially stems from antimicrobial resistance (AMR), which is a microorganism’s ability to withstand antimicrobial agents. This can stem from natural resistance, which is when specific bacterial genera or species have unique structural or functional characteristics that make them resistant to certain antibiotics, or acquired resistance, which is when naturally susceptible bacteria develop antibiotic resistance by mutating or by horizontally acquiring resistance genes. One approach used to address this issue is Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS). This technique is used to identify and analyze biomolecules, especially proteins and peptides, and is used in various research areas, clinical diagnostics, and industrial applications due to its high sensitivity, speed, and versatility. With AMR, MALDI-TOF can help identify subtle proteomic difference that determine whether a spectrum corresponds to a sample susceptible or resistant to a particular antibiotic.
In this review article, the scientists aimed to evaluate the current state of the art in using machine learning for detecting and classifying AMR from MALDI-TOF-MS data. Forty studies were involved, with Staphylococcus aureus, Klebsiella pneumoniae and Escherichia coli being the most frequently cited bacteria. The most studied incidents of antibiotics resistance corresponded to methicillin for S. aureus, cephalosporins for K. pneumoniae, and aminoglycosides for E. coli. Further, the most employed algorithms for predicting AMR were random forest, support vector machine and logistic regression. Seven studies were also reported using artificial neural networks. Most of the studies involved reported metrics such as accuracy, sensitivity, specificity, and the area under the receiver operating characteristic (AUROC) above 0.80.
Altogether, the review showed that random forest, support vector machine, and logistic regression are effective for predicting antimicrobial resistance using MALDI-TOF MS data. Additionally, deep learning techniques have potential in this area. For future research, deep learning and multi-label supervised learning should be explored further for comprehensive antibiotic resistance prediction in clinical practice.
References
(1) López-Cortés, X. A.; Manríquez-Troncoso, J. M.; Kandalaft-Letelier, J.; Cuadros-Orellana, S. Machine Learning and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectra for Antimicrobial Resistance Prediction: A Systematic Review of Recent Advancements and Future Development. J. Chromatogr. A 2024, 1734, 465262. DOI:
(2) Antibiotic Resistance. Cleveland Clinic 2024.
Newsletter
Join the global community of analytical scientists who trust LCGC for insights on the latest techniques, trends, and expert solutions in chromatography.