Kinase Fragments Dimerize Without Oligomerization Domains, Shown by SEC-MALS
The Application Notebook
etermination of oligomeric states is an important issue in protein chemistry. For example, self-assembly via oligomerization domains is crucial for the regulation of several protein kinases. Determination of the oligomeric state of fragments of these kinases is a means of verifying the involvement of each domain in self-assembly.
Determination of oligomeric states is an important issue in protein chemistry. For example, self-assembly via oligomerization domains is crucial for the regulation of several protein kinases. Determination of the oligomeric state of fragments of these kinases is a means of verifying the involvement of each domain in self-assembly.
Analytical size-exclusion chromatography (SEC) is widely used for determining molar mass and oligomeric state of proteins in solution, but it exhibits some important limitations. For example, interactions of proteins with column material can lead to delayed elution and hence erroneous results when relying on column calibration. Since even ideal elution occurs according to hydrodynamic size rather than true molecular weight, there are no appropriate molar mass gel filtration standards for analysis of proteins, fragments, or complexes of non-globular structure that present a different size or molecular weight dependence than globular proteins.
The use of size-exclusion chromatography in combination with multi-angle light scattering (SEC-MALS) determines molecular weights independently of elution time and conformation, overcoming the need for standards and the errors inherent in analytical SEC. In SEC-MALS, chromatography is used solely to separate the individual species so they can be characterized by light scattering for biophysical properties such as molar mass, size, conformation, and degree of conjugation.
This note describes the analysis of a kinase fragment lacking its association domain in order to determine its oligomeric state in solution. SEC-MALS revealed that the kinase moiety clearly remains dimeric in solution, even in the absence of its purported oligomerization domain.
Experimental Conditions
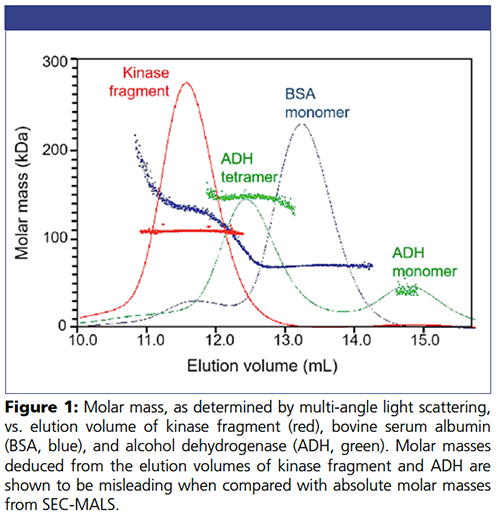
An HP-SEC column was calibrated using bovine serum albumin (BSA) monomer and dimer. The kinase fragment and alcohol dehydrogenase (ADH, 38 kDa monomer and 150 kDa tetramer) were each run on the column, and the elution times compared to those of BSA monomer (66.4 kDa) and dimer (133 kDa). Absolute molar mass (MW) of the proteins at each elution volume were determined by analysis of signals from the multi-angle light scattering and refractive index detectors (DAWN and Optilab, respectively) in ASTRA software. Chromatograms were overlaid with molecular weight values calculated for each elution time along the peaks, as seen in Figure 1.
Results and Discussion
The monomeric kinase fragment has a sequence molar mass of 53.5 kDa. The fragment (red trace) eluted at nearly the same volume as the BSA dimer (blue trace), suggesting that its molar mass is approximately 140 kDa, or trimeric. However, MALS determines an absolute molar mass in solution of 108 kDa, revealing that the protein is actually a dimer. The molecular weight is absolutely uniform across the peak, indicating a high degree of homogeneity. Such early elution is indicative of a non-globular conformation.
ADH tetramer (green trace, 150 kDa) eluted between the monomer and dimer of BSA, possibly because of ADH-column interactions that caused it to elute late relative to its size. Comparison of the fragment’s elution volume to ADH monomer and tetramer would mislead the investigator to assume a tetrameric state, even further removed from the truth than comparison to BSA.
Conclusions
SEC-MALS provides true solution molecular weight for proteins, overcoming the inherent errors produced by reliance on column calibration. Here we have shown by SEC-MALS that kinase fragments are dimeric, even without the purported oligomerization domain; but they are not trimeric or tetrameric as might have been deduced via column calibration. The addition of a DAWN MALS detector to standard SEC reveals the essential biophysical properties of proteins, fragments, and complexes.

Wyatt Technology Corp.
6330 Hollister Ave, Santa Barbara, California 93110, USA
Tel.: 1 (805) 681 9009
Website: www.wyatt.com

MALDI Guided SpatialOMx® Uncovers Proteomic Profiles in Tumor Subpopulations of Breast Cancer
September 1st 2020The timsTOF fleX system bridges a current gap by providing MALDI Imaging and in-depth proteomics analysis in just one instrument. The instrument offers all benefits of a timsTOF Pro for time-efficient and sensitive proteomics, combined with a high-resolution MALDI source and stage. Using PASEF technology, it is possible to retrieve high protein ID rates with small sample amounts. Here we present the new SpatialOMx® workflow to identify distinct proteomic profiles for different tumor subpopulations in breast cancer as an example for this powerful approach.