The Role of Ion Pairing Agents in Liquid Chromatography (LC) Separations
Ion pairing agents are frequently used in liquid chromatography (LC), especially in reversed-phase liquid chromatography (RPLC), to increase the separation and retention of charged analytes. However, their use in other modes of HPLC, such as sizeexclusion HPLC (SE–HPLC) and ion exchange chromatography (IEC) has not been extensively explored. In RPLC, the target analytes may have charged functional groups, which makes it difficult to maintain them on the hydrophobic stationary phase. This difficulty is often overcome by the inclusion of an ion pairing agent, which introduces ionic contacts and strengthens the hydrophobic connections between the analytes and stationary phase. The separation mechanisms in both SE–HPLC and IEC rely on size and charge differences, respectively, between analytes. Thus, they are naturally constructed to separate analytes without the requiring extra ion pairing agents. In this study, we introduced an ion pairing agent (<0.1% formic acid) to the protein sample, and as a result, an alteration in the elution profile (separation efficiency, selectivity, retention of analytes, and resolution) has been achieved. When samples were introduced with an ion-pairing agent in SE–HPLC, there was a considerable improvement in low molecular weight species (LMWs) separation with four extra peaks with higher resolution (up to 2%), without significant alteration in total area percentage. Similarly, a minor new acidic variant peak was resolved in the weak cation-exchange LC (WCX–LC) analysis of the formic acid-enhanced sample, with the resolution being increased by 3%. The results show that using ion pairing agents should be explored in other modes of HPLC as well as for applications where they may offer enhanced chromatographic separations.
Reversed-phase liquid chromatography (RPLC) is a simple and user-friendly technology that is often used for separation of bioanalytes, thereby accounting for up to 70–80% of bioseparations (1). Most acidic samples have less ionization and would be effectively retained under these conditions, along with neutral molecules. Basic analytes would ionize, but if the majority of the sample molecule is nonpolar, sufficient retention is achieved (2). Many basic substances will not ionize at pH 10, allowing them to undergo chromatographic separation. However, other bases will still ionize at this pH, resulting in poor retention (3). Therefore, charged functional groups of target analytes make it challenging to retain them on a hydrophobic stationary phase, particularly in RPLC.
Often, an ion pairing agent is used to alleviate this problem (4). Because broad, trailing peaks are difficult to accurately quantify, an ion pairing agent can be used to enhance peak shape, and as a result, they produce peaks that are sharper and symmetric, resulting in improved accuracy and precision (5). Protein separation selectivity can also be increased by using an ion pairing agent, facilitating improved resolution between closely related protein species by differentiating proteins with comparable hydrophobic features and charge traits. When examining protein variations or alterations, complicated protein combinations, or both, this is especially useful (6).
To improve the separation of ionizable compounds, LC employs a number of ionpairing agents. Alkylamines are frequently used as ion pairing agents to enhance the separation of ionizable substances. They interact with negatively charged analytes through electrostatic attractions and serve as cationic ion pairing agents. Examples include tetraalkylammonium salts, such as tetrabutylammonium phosphate and tetraethylammonium bromide (TBAP) (7). Alkylsulfonates are anionic ion pairing substances that work with analytes that are positively charged through electrostatic attractions. For ion pairing in LC, sodium dodecylsulfate (SDS), a popular alkylsulfonate, is used for the same (8). In LC, perfluorinated carboxylic acids such as heptafluorobutyric acid (HFBA) and nonafluoropentanoic acid (NFPA) are frequently utilized as ion pairing agents. They work as anionic ion pairing agents and interact with cationic analytes. Cationic analytes interact with perfluorinated carboxylic acids, which balances their charge. Perfluorinated carboxylic acids make it easier for the analyte to remain on the stationary phase of the LC column by neutralizing the charge (9). Quaternary ammonium compounds, such as acetyltrimethylammonium bromide (CTAB) and tetrabutylammonium bromide (TBAB), interact with negatively charged analytes as cationic ion pairing agents. They are frequently employed in RPLC to enhance analyte separation (10). The separation of basic substances can be accomplished with the help of perfluorinated alkylsulfonates, which are anionic ion pairing agents with fluorinated alkyl chains. In this category, perfluorobutanesulfonic acid (PFBS) and perfluorooctanesulfonic acid (PFOS) are frequently utilized (11). For the separation of metal ions, crown ethers are utilized as selective ion pairing agents with examples, including 18-crown-6, dibenzo-18-crown-6, and benzo-15-crown-5. These work by creating stable complexes with the metal ions. A cyclic arrangement of oxygen atoms is part of the peculiar structure of crown ethers. These oxygen atoms are capable of building stable complexes by coordinating with the metal ions. The metal ion is successfully contained within the crown ether cavity, which isolates it from its surroundings. Depending on the size and charge of the metal ion and the size of the crown ether cavity, this complexation is extremely selective (12). It is important to remember that the pKa, charge, and desired separation conditions all affect the ion pairing agent that is used. To achieve the best separation, the concentration of the ion pairing agent in the mobile phase is also very important. It’s also critical to take into account how well the ion pairing agent works with the LC system and the chosen detection strategy.
As an ion pairing agent in LC, formic acid is easily accessible and often employed. It is readily available for commercial use from many chemical vendors in a high purity, making it convenient for use in laboratories. Regulatory organizations, including the European Food Safety Authority (EFSA) and the U.S. Food and Drug Administration (FDA), recognize formic acid as generally recognized as safe (GRAS) (13). As an addition or preservative, it is widely utilized in the food and beverage sectors. Because of its well-known safety profile, it can be used in analytical laboratories with little to no regulatory interference (14). When properly stored in sealed containers, formic acid offers excellent stability. Its reasonably long shelf life enables prolonged usage without noticeably degrading or losing performance. This makes it convenient for long-term storage and usage in laboratories. Its adaptability to multiple LC systems gives it a flexible and affordable option for ion pairing in a range of analytical applications (15,16).
Keeping the above in mind in this study we demonstrated the impact of selective ion pairing agent in LC separation. We have integrated up to 0.1% formic acid in the samples before injection and observed significant improvement in low molecular weight species (LMWs) separation with four additional small peaks, with higher resolution on the LMWs (2%). In the WCX–LC separation, the formic acidintegrated sample outperformed the nonintegrated sample in terms of major peak resolution and separated on extra minor acidic variant. As a result, it suggests that depending on the application at hand, ion pairing agents may be able to enhance chromatographic separations.
Materials and Methods
A humanized IgG1 kappa expressed inhouse monoclonal antibody (mAb) tratuzumab was used in the investigation. Prior to analysis, sample concentration was determined using ultraviolet (UV) absorbance at 280 nm and corrected to 2 mg/mL. The size-exclusion chromatography (SEC) and WCX elution profiles of the research sample were evaluated both with and without the addition of an ion pairing agent (up to 0.1% formic acid) to evaluate the intrinsic impact of the agent in the sample. For the evaluation under research, a standard monoclonal antibody (mAb) sample for the WCX experiment and a stress mAb sample (kept in room temperature for 4 h) for the SEC experiment were used.
Case Study 1: WCX Chromatography
The BiomAb NP5, PK 5 µm nonporous (4.6 x 250 mm, Agilent Technologies) column was used to separate charge variations of the sample under examination. Ammonium acetate (50 mM) was used to make the buffers, with a pH of 6.8 as buffer A and 10.11 as buffer B, respectively. Separation was conducted through a step gradient with a linear increase in %B up to 56% until 8.60 min followed by an increase of %B from 56% (8.60 min) to 70% (11.00 min), with the temperature being maintained at 30 °C. Further washing and equilibration was performed from 11.10 to 13 min at 100% B followed by 100% B up until 15 min. The UV absorption signal was monitored at 280 and 220 nm, respectively. Comparison of non-volatile buffer was performed (150 mM sodium phosphate buffer and 0.05% NaN3 at pH 6.8 as buffer A and 150 mM sodium phosphate buffer and 0.05% NaN3 at pH 10.11 as buffer B) and insignificant deviation was observed (p-value higher than 0.05) compared to the volatile buffer (data not shown). A standard trastuzumab sample with and without formic acid (0.1%) was used to evaluate the impact of ion painting agent in LC separation while integrated with sample just before the injection was done.
Case Study 2: SE–HPLC Chromatography
To quantify aggregates present in the stressed sample (Trastuzumab, IgG1, in room temperature for 4 h), size-exclusion high performance LC (SE-HPLC) was carried out on an AdvanceBio SEC 200A 1.9 mm 4.6 x 300 mm (Agilent Technologies) column run on the Dionex Ultimate 3000 ultrahigh-pressure LC (UHPLC) system (Thermo Fisher Scientific) at 25 °C, with and without <0.1% formic acid prior to injection. After loading 50 µL of sample at 2 mg/mL, isocratic elution was performed for 15 min at a flow rate of 0.5 mL/min, using a solution of 50 mM sodium phosphate buffer, 300 mM NaCl, and 0.05% NaN3 at pH 6.8. Before usage, all buffers underwent degassing and filtration using a 0.2 mm cut-off polyethersulfone membrane filter (Pall Corporation). Chromeleon software from Thermo Fisher Scientific was used for data analysis. UV absorbance at 280 nm was monitored for detection.
Results and Discussion
Case Study 1: Impact of Ion Pairing Agent in WCX Chromatography
It was observed that the sample integrated with 0.1% formic acid was resolved into 10 consecutive peaks compared to sample without an ion pairing agent, which resolved the sample into nine peaks after WCX in the LC separation. The total acidic area in both with and without formic acid-integrated samples were found to be 32.67% and 32.99%, with six and five acidic peaks separated, respectively. It is evident from Table I that the peak separated at a retention time (t R) of 6.23 min in formic acidintegrated sample has not been resolved in sample without formic acid (Figure 1). The formic acid-integrated sample resolution was higher (53.50%) compared to the without formic acid sample (50.67%). It also had the same injection volume (50 µL), and elution occurred approximately at the same retention time (13.25 min and 13.24 min, respectively). In the case of postpeak analysis, the sample with formic acid was found to be resolved into three basic variants, similar to a sample without formic acid, which also resolved into three basic variants. However, the total area percentage of the basic variants was found to be 13.83% compared to 16.34% for sample without formic acid.
Table I: Identification and quantification of peaks obtained from WCX chromatographic separation of IgG1 (trastuzumab), with and without integrated 0.1% formic acid (FA).
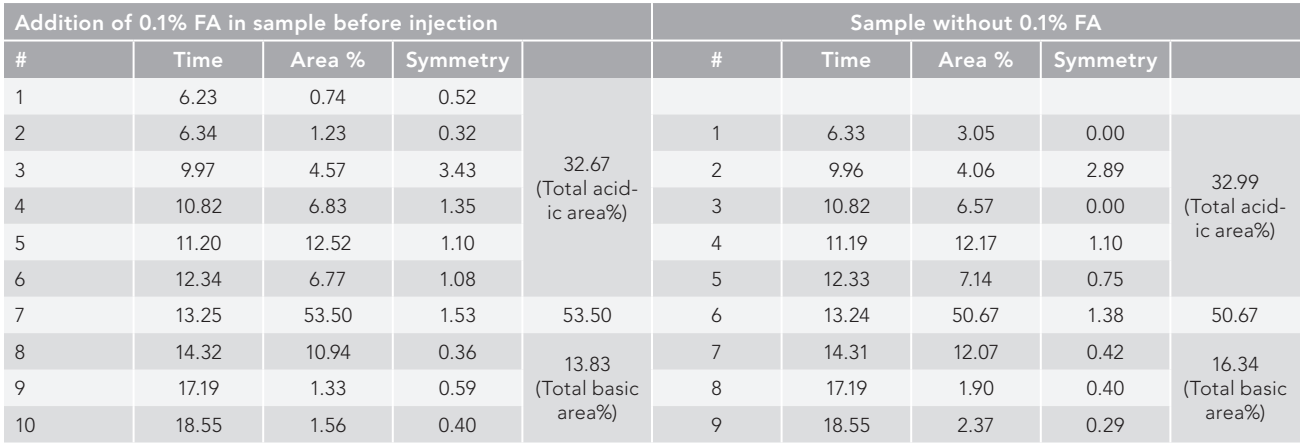
Figure 1: Peaks identified from WCX chromatographic separation of IgG1 (trastuzumab), (a) with and (b) without integrated 0.1% formic acid. The sample integrated with 0.1% formic acid was resolved into 10 consecutive peaks compared to the sample without an ion pairing agent, which resolved the sample into nine peaks after WCX in LC-separation.
In contrast to the sample without an ion pairing agent, which resolved the sample into nine peaks following WCX in LC-separation, the sample integrated with 0.1% formic acid was resolved into 10 continuous peaks. The overall acidic area, divided into six and five acidic peaks, was found to be 32.67% and 32.99%, respectively, in samples with and without formic acid integrated. The peak that was separated in tR 3.23 min in the formic acid-integrated sample was not resolved in the sample without formic acid, as shown in Table I (Figure 1). With the same injection volume (50 µL) and roughly the same retention time, the formic acid (FA)-integrated sample resolution was greater (53.50%) than it was without the FA sample (50.67%) at 13.27 min and 13.24 min, respectively. In comparison to the norm, the resolution of the main peak was found to be 3% greater in the formic acid-integrated sample (Table I). It was observed during post-peak analysis that the sample with formic acid resolved into three basic variants, just like the sample without formic acid, which did the same. In comparison to the sample lacking formic acid, the overall basic area percent of fundamental variations was found to be greater at 16.34% compared to 13.83% for the sample with formic acid.
One additional peak (tR, 6.32 min) with 0.74% was found in acidic variants when comparing the formic acid-integrated and non-integrated profiles of WCX-LC separation in HPLC. This peak can be distinguished in high-sensitive instruments like mass spectrometry (MS) total ion chromatogram (TIC) profiles (17), but not in HPLC separation (18), which explains why integration of ion pairing agents increases resolution and sensitivity when compared to non-integrated samples.
Case Study 2: Impact of Ion Pairing Agent in LC Separation for SE–HPLC Chromatography
SE–HPLC chromatograms for samples with and without an ion pairing agent (<0.1% formic acid) are presented in Table II, and it is seen that better resolution was achieved (nine peaks separated) when integrated with 0.1% formic acid than in absence of formic acid sample (five peaks separated). Total high molecular weight (HMW) area percentage and the number of HMW peaks were found to be identical when comparing integrated formic acid sample with nonintegrated sample, coming in at 23.68% and 23.59% with three acidic peaks in both, respectively. The variations, however noticed, in LMW peak separation, and resolved into five successive peaks (19) and one peak (20), for formic acid-integrated and nonintegrated samples, keeping the total LMW area of 8.91% and 7.46%, respectively (Figure 2).
Table II: Identification and quantification of peaks resolved in SE-HPLC separation of RT stress IgG1 (trastuzumab), with and without integrated <0.1% (formic acid [FA]) prior to injection.
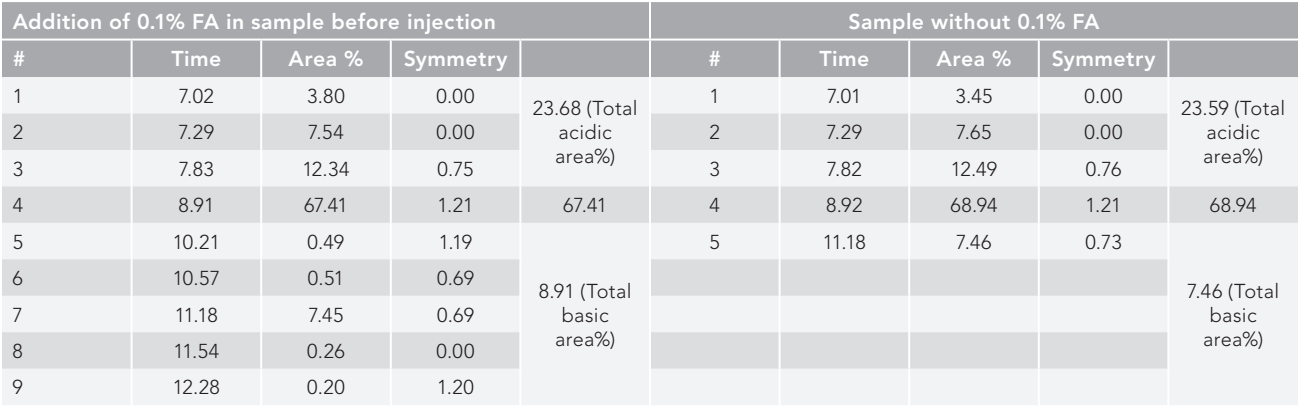
Figure 2: SE-HPLC separation and identification of peaks of RT stress IgG1 (trastuzumab), (a) with and (b) without integrated <0.1% formic acid prior to injection. The stress sample under study resolved better (split into nine peaks) when integrated with 0.1% formic acid than when the formic acid sample was not present (separated into five peaks).
Conclusion
Ion pairing agents are commonly used in reversed-phase liquid chromatography (RPLC) to increase the separation and retention of charged analytes. However, as we demonstrate in this article, using ion pair agents could be extended to other modes of HPLC, such as size exclusion (SE–HPLC) and ion exchange chromatography (IEC). In this study, considerable enhancement in separation of low molecular weight species (LMWs) was observed with higher resolution (up to 2%) without altering the total area percentage. Additionally, in the WCX–LC separation, the formic acid-integrated sample yielded significantly better resolution of the main peak and resolved into one additional minor acidic variations than the non-integrated sample.
References
(1) Dolan, J. Ion pairing—blessing or curse?. LCGC Eur. 2008, 21 (5), 258–263.
(2) Ståhlberg, J. Retention models for ions in chromatography. J. Chromatogr. A. 1999, 855 (1), 3–55.
(3) Liu, A., Cheng, M., Zhou, Y.; et al. Bioanalysis of Oligonucleotide by LC–MS: Effects of Ion Pairing Regents and Recent Advances in Ion-Pairing-Free Analytical Strategies. Int. J. Mol. Sci. 2022, 23 (24), 15474.
(4) Noh, G.; Keum, T.; Bashyal, S.; et al. Recent progress in hydrophobic ion-pairing and lipid-based drug delivery systems for enhanced oral delivery of biopharmaceuticals. J. Pharma. Investig. 2022, 52 (1), 1–19.
(5) McCalley, D. Understanding and managing peak shape for basic solutes in reversed-phase high performance liquid chromatography. Chem. Commun. 2023, 59, 7887–7899.
(6) Gussakovsky, D.; Anderson, G.; Spicer, V.; et al. Peptide separation selectivity in proteomics LC‐MS experiments: Comparison of formic and mixed formic/heptafluorobutyric acids ion‐pairing modifiers. J. Sep. Sci. 2020, 43 (20), 3830–3839.
(7) Donegan, M.; Nguyen, J. M.; Gilar, M. Effect of ion-pairing reagent hydrophobicity on liquid chromatography and mass spectrometry analysis of oligonucleotides. J. Chromatogr. A. 2022, 1666, 462860.
(8) Wibel, R.; Knoll, P.; Le-Vinh, B.; Kali, G.; Bernkop-Schnürch, A. Synthesis and evaluation of sulfosuccinate-based surfactants as counterions for hydrophobic ion pairing. Acta Biomater. 2022, 144, 54–66.
(9) Kotrebai, M.; Tyson, J.F.; Block, E; Uden, P. C. High-performance liquid chromatography of selenium compounds utilizing perfluorinated carboxylic acid ion-pairing agents and inductively coupled plasma and electrospray ionization mass spectrometric detection. J. Chromatogr. A. 2000, 866 (1), 51–63.
(10) Tang, H.; Geng, K.; Hao, J.; et al. Properties and stability of quaternary ammonium-biphosphate ion-pair poly (sulfone) s high temperature proton exchange membranes for H2/O2 fuel cells. J. Power Sources 2020, 475, 228521.
(11) García-Alvarez-Coque, M. C.; Torres-Lapasió, J.R.; Ruiz-Angel, M. J.; et al. Secondary chemical equilibria in reversed-phase liquid chromatography. In Liquid Chromatography; Elsevier, 2023; pp 121–143.
(12) Zaleskaya-Hernik, M.; Karbarz, M.; Romański, J. The use of microelectrodes to study ion recognition by a squaramide-based ion pair receptor consisting of a ferrocene reporter. J. Electroanal. Chem. 2023, 928, 117058.
(13) Cai, W.; Tang, F.; Zhao, X.; et al. Different lactic acid bacteria strains affecting the flavor profile of fermented jujube juice. J. Food Process. Preserv. 2019, 43 (9), e14095.
(14) Yadav, M.; Dhyani, S.; Joshi, P.; et al. Formic acid, an organic acid food preservative, induces viable-but-non-culturable state, and triggers new Antimicrobial Resistance traits in Acinetobacter baumannii and Klebsiella pneumoniae. Front. Microbiol. 2022, 13, 966207.
(15) Yang, J.; Marzan, T. A.; Ye, W.; et al. A cautionary tale: quantitative LC-HRMS analytical procedures for the analysis of N-nitrosodimethylamine in metformin. The AAPS Journal 2020, 22, 1–8.
(16) Ma, W.; Yang, B.; Li, J.; et al. Development of a simple, underivatized method for rapid determination of free amino acids in honey using dilute-and-shoot strategy and liquid chromatography-tandem mass spectrometry. Molecules 2022, 27 (3), 1056.
(17) Bailey, A. O., Han, G., Phung, W., et al. Charge variant native mass spectrometry benefits mass precision and dynamic range of monoclonal antibody intact mass analysis. MAbs 2018, 10 (8), 1214–1225. DOI: 10.1080/19420862.2018.1521131
(18) Rea, J. C.; Wang, Y. J.; Moreno, T. G.; et al. Monoclonal antibody development and physicochemical characterization by high performance ion exchange chromatography. Innovations in biotechnology. InTech 2012, 439–464.
(19) Lu, X., Machiesky, L. A., De Mel, N., et al. Characterization of IgG1 Fc deamidation at asparagine 325 and its impact on antibody-dependent cell-mediated cytotoxicity and FcγRIIIa binding. Sci. Rep. 2020, 10 (1), 1–11.
(20) Yang, R.; Tang, Y.; Zhang, B.; et al. High resolution separation of recombinant monoclonal antibodies by size-exclusion ultra-high performance liquid chromatography (SE-UHPLC). J. Pharm. Biomed. Anal. 2015, 109, 52–61.
About the Co-Authors
Anurag S. Rathore is a professor in the Department of Chemical Engineering at the Indian Institute of Technology in Delhi, India.

Sanghati Bhattacharya is a post doctoral fellow with Prof. Anurag S. Rathore at the COE for Biopharmaceutical Techonology, IIT Delhi, India.

About the Column Editor
Jared Auclair is an Associate Dean of Professional Programs and Graduate Affairs at the College of Science at Northeastern University, in Boston, Massachusetts. He is also the Director of Biotechnology and Informatics, as well as the Director of the Biopharmaceutical Analysis Training Laboratory. Direct correspondence to: LCGCedit@mmhgroup.com


Supercritical Fluid Chromatography for Separating Chemotherapy Anti-Nausea Medication
October 30th 2024Scientists from Peking Union Medical College & Chinese Academy of Medical Sciences created a new method for separating palonosetron hydrochloride, which is frequently used to prevent acute or delated nausea and vomiting caused by chemotherapy.
HILIC Peptide Retention Times Predicted Using New Approach
October 29th 2024Manitoba Centre for Proteomics and Systems Biology scientists produced a new means of predicting peptide retention times for hydrophilic interaction liquid chromatography (HILIC) at acidic pH in formic-acid based eluents.