The Role of Liquid Chromatography–Tandem Mass Spectrometry-Based Metabolomics in Disease Biology
Metabolomics is the study of metabolites present in a biological system. Liquid chromatography tandem mass spectrometry (LC–MS/MS)-based metabolites typically use multiple high performance liquid chromatography (HPLC) columns with different selectivity in a single run to maximize metabolite separation and improve overall metabolite coverage. The selection of an appropriate HPLC column for an LC–MS/MS-based metabolite depends on several factors, including the physicochemical properties of the metabolite of interest, matrix complexity, and the sensitivity and selectivity required for the analysis.
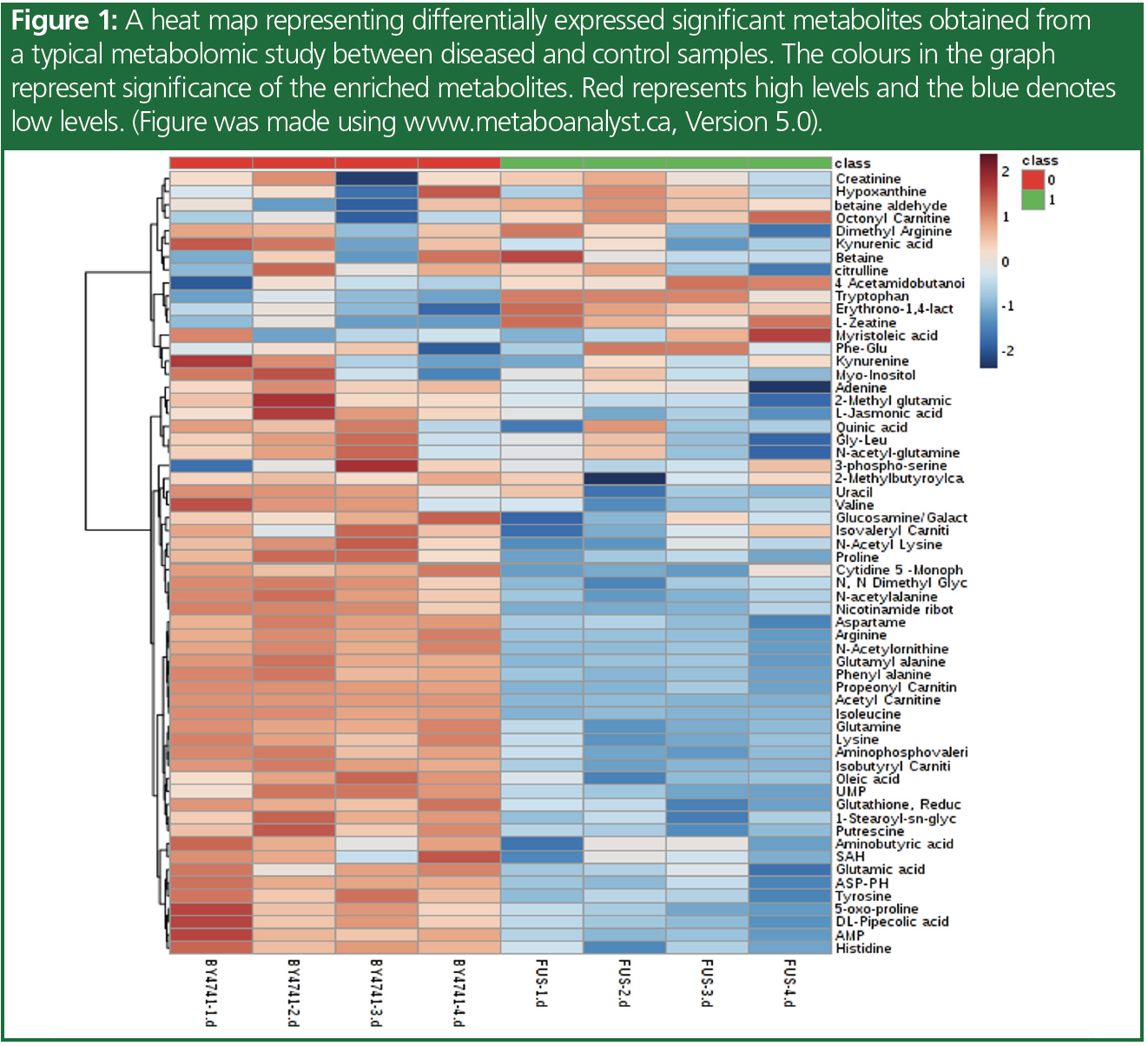
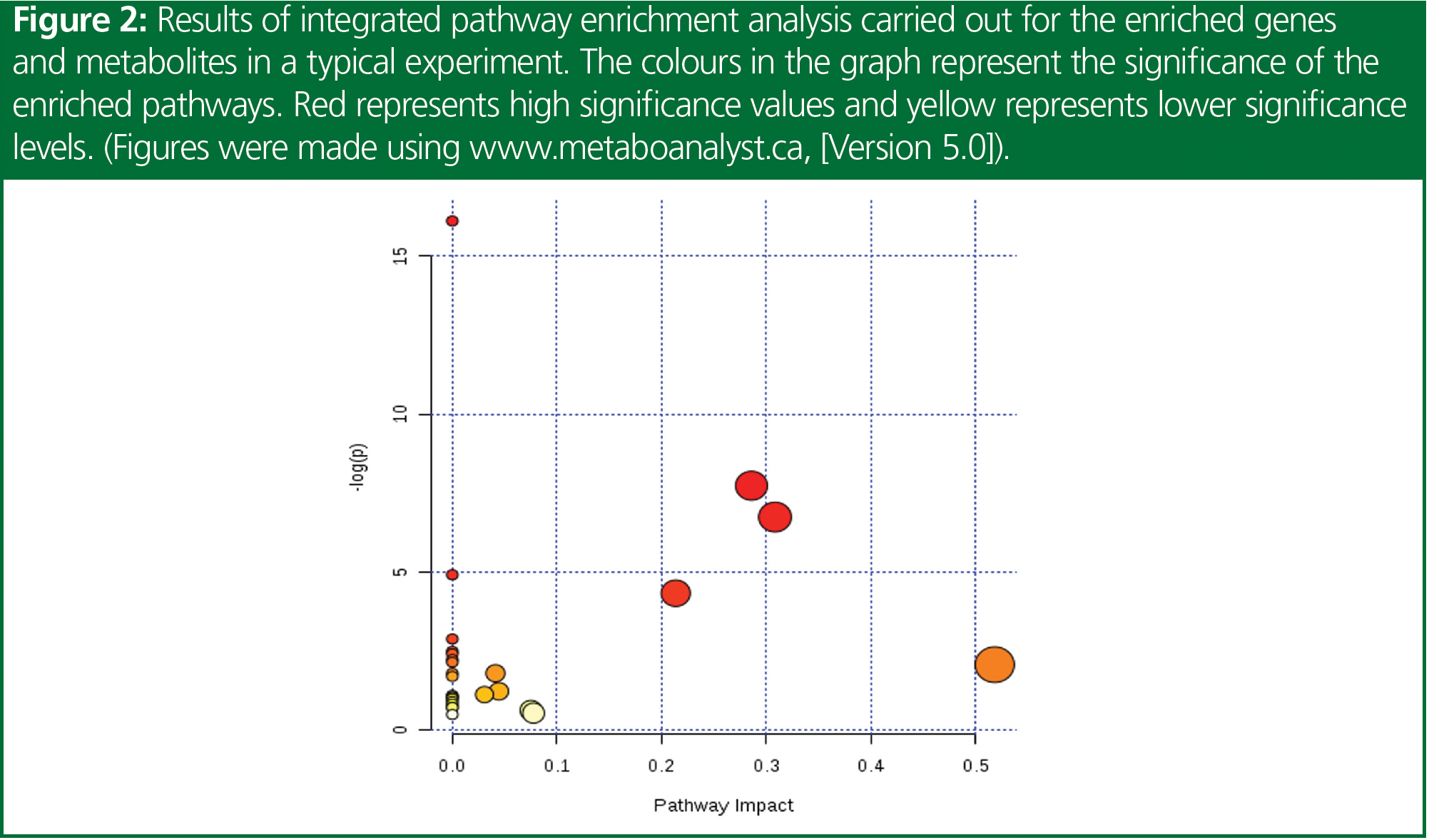
Metabolites are small molecules produced by the metabolic pathways in the body, and their levels can be influenced by genetic and environmental factors. Metabolomics is the study of the metabolites present in a biological system, which is increasingly gaining importance in the field of disease biology. It is a comprehensive approach that aims to identify and quantify all metabolites present in a biological system, and provides insights into the underlying biochemical processes. The metabolome represents the cumulative change that is observed at the level of genome, epigenome, transcriptome, proteome, and their regulation, and hence are very near to the phenotype. Figure 1 represents a heat map for the significant metabolites that are observed in a sample study. Understanding metabolic changes, or the deregulated pathway therein, could help to explain the disease phenotype, associated mechanisms, potential biomarkers for progression and prognosis, as well as potential therapeutic targets. Figure 2 represents a pathway enrichment study to identify the different pathways involved or affected to further understand the metabolic changes occurring.
Liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS) is a powerful technique that has been widely used in metabolomics research. This technique separates metabolites based on their physicochemical properties and detects them with high sensitivity and specificity using mass spectrometry.
In cancer research, LC–MS/MS-based metabolomics has been used to identify metabolites that are differentially expressed between cancer and non-cancer cells or tissues. In clinical diagnostic testing, these metabolites can serve as biomarkers for cancer diagnosis and prognosis, as well as potential targets for therapy. For example, the levels of certain amino acids, such as serine and glycine, are often elevated in cancer cells, which could be exploited for therapeutic purposes. Cancers also utilize glutaminolysis and inhibitors of glutaminase help in better prognosis. Further, in a cancer microenvironment, immune cells do not recognize cancer cells as a threat and the role of adenosine signalling is monitored in the process.
Taken together these results show that metabolites from deregulated metabolic pathways could be potential biomarkers and therapeutic targets in cancer (1).
In diabetes research, LC–MS/MS has been used to identify metabolic pathways that are dysregulated. This information can then be used further to develop new drugs or dietary interventions that target these pathways to improve glucose control and prevent complications associated with diabetes. For example, LC–MS/MS-based metabolomics has identified several metabolites associated with insulin resistance, such as branched‑chain amino acids and acylcarnitines, which could be used as therapeutic targets (2).
In cardiovascular disease research, LC–MS/MS-based metabolomics has been used to identify biomarkers to help understand the risk of cardiovascular events, such as heart attack, heart failure, and stroke. For example, the levels of certain lipids, such as phosphatidylcholine and sphingomyelin, have been shown to be associated with cardiovascular risk, and could be used to develop personalized preventive strategies. Further, homocysteine and dimethyl arginine are shown to be risk factors for many diseases including cardiovascular diseases. Homocysteine leads to inflammation, mitochondrial dysfunction, and lipid and cholesterol biosynthesis. In some research studies, homocysteine reduction therapy using vitamin B12 has shown diverse effects (3).
LC–MS/MS-based metabolomics has been used to identify metabolic pathways that are altered for neurodegenerative diseases, such as Alzheimer’s (AD), Parkinson’s, Huntington’s Disease (HD), and amyotrophic lateral sclerosis (ALS). Studies conducted have shown an association of deregulated metabolic pathways with HD. Interestingly, metabolites from these deregulated pathways modulated aggregation of mutant HD (4). Further, analysis of transcriptomic and proteomic data sets showed low expression of B12 and folate transporters. Studies have shown low expression levels as well as activity of pyridoxal kinase in HD model systems. Since vitamins like B6, B12, and folate act as cofactors in various enzymatic reactions, an analysis of a cofactor-enzyme/protein interaction network was generated and the proteins were grouped into pathways (5). The metabolic pathways that require B6, B12, and folate showed considerable overlap, with deregulated metabolic pathways in HD, as well as helping to clear mutant Huntington’s aggregates in a yeast model HD (5).
Metabolomic studies on a yeast model of ALS showed an association of deregulated metabolic pathways like glutathione and fatty acids, as well as nicotinate and nicotinamide pathway with disease. Reduced glutathione, short-chain fatty acids, and nicotinate mitigated aggregation of mutant TDP43 in a yeast model of ALS, while long‑chain fatty acids exacerbated it (6).
These studies demonstrate the potential of targeting metabolic pathways by dietary interventions to alter the course of the diseases. Further, studies in neurodegenerative disease such as HD and AD have shown that atrophy of the brain, volume, and metabolic changes precede the onset of symptoms (7,8). In light of these findings, maintaining metabolic homeostasis could help delay onset of disease and aid in better prognosis (7).
Metabolomic analysis has been performed on different forms of glaucoma. Studies have shown changes in the levels of amino acids and fatty acids. Our studies have demonstrated an association of immune‑metabolism and activation of tryptophan metabolism in primary open angle glaucoma (9). Further, studies have shown deregulation of nicotinate and nicotinamide metabolism in glaucoma. These results, concomitant with elevated levels of cytokines, show a potential role for inflammation and immunometabolism with disease progression.
HPLC Columns and Column Selection for Metabolomics
High performance liquid chromatography (HPLC) column selectivity plays an important role in LC–MS/MS-based metabolomics. This is because it determines the extent to which metabolites are separated and detected for research. Selectivity refers to the ability of a column to separate analytes based on physical and chemical properties, such as size, polarity, and charge. LC–MS/MS-based metabolomics often use multiple HPLC columns with different selectivities in a single run to maximize metabolite separation and improve overall metabolome coverage. This allows for a more comprehensive analysis of metabolites present in biological samples.
For example, reversed-phase HPLC columns are widely used in LC–MS/MS-based metabolomics because they can separate metabolites based on their hydrophobicity. This is particularly useful for separating lipids, which are highly hydrophobic and often difficult to separate with other column types. However, reversed-phase HPLC columns may not resolve all metabolites in a sample, especially metabolites with similar hydrophobicity. In such cases, other types of HPLC columns with different selectivity, such as hydrophilic interaction liquid chromatography (HILIC) or ion exchange chromatography (IEC), can be used to separate the remaining metabolites.
The selection of a suitable HPLC column for LC–MS/MS-based metabolomics depends on several factors, including the physicochemical properties of the metabolites of interest, the complexity of the sample matrix, and the analytical sensitivity and selectivity required for the analysis. Here are some of the general steps to follow to choose a suitable HPLC column for metabolomics.
First, determine the physicochemical properties of the metabolite of interest. Metabolite properties such as size, polarity, charge, and hydrophobicity influence the choice of HPLC column. In general, reversed-phase columns are commonly used for hydrophobic metabolites, whereas HILIC columns are more useful for polar metabolites. The composition of the biological matrix in which metabolites reside, also referred to as the sample matrix, can affect the separation efficiency and selectivity of the column. For example, biological samples such as plasma and urine are complex matrices containing high concentrations of interfering compounds that can affect analytical sensitivity and selectivity. In such cases, a high-resolution column with improved selectivity can be used for efficiency gains, or the proper sample preparation must be employed prior to separation analysis.
Next, evaluate the required analytical sensitivity and selectivity of the analysis. These parameters are also influenced by the choice of LC column. For example, if the metabolite of interest is present at low concentrations, sensitive columns using advanced particle chemistry can be used. Alternatively, if the metabolite of interest is structurally similar to that of other metabolites, a chiral column can be used. Once the column has been selected, chromatographic conditions such as mobile phase composition, pH, and temperature should be optimized to achieve optimal separation and detection of metabolites.
Below are some examples of how different types of HPLC columns are used in metabolomics research.
Reversed-phase HPLC columns are widely used in lipidomics research to separate and identify lipid species present in biological samples. They were used to separate and identify different lipid species in the brain tissue of rats with AD, demonstrating significant alterations in lipid metabolism in this disease (10).
Ion-exchange HPLC columns are used to separate and identify charged metabolites such as amino acids, peptides, and nucleotides. HILIC columns are commonly used to separate polar and hydrophilic metabolites such as sugars, amino acids, and nucleotides. HILIC columns have been used to separate and identify polar metabolites present in urine samples to display significant alterations in the levels of certain metabolites for specific diseases, such as multiple sclerosis (MS) (11).
Size-exclusion chromatography (SEC) columns are commonly used to separate molecules based on their size, making them particularly useful for the analysis of large biomolecules such as proteins and nucleic acids. For example, SEC–HPLC has been used to separate and identify peptides and proteins in biological samples such as cerebrospinal fluid (CSF). A study identified several proteins that were differentially expressed in the CSF of Alzheimer’s samples compared to controls, providing insights into the underlying pathology of the disease (12).
In summary, LC–MS/MS-based metabolomics is a widely used and powerful tool in the study of disease biology. This technique enables the identification and quantification of metabolites in biological systems, providing insight into underlying biochemical processes. By using LC–MS/MS-based metabolomics, scientists and researchers have identified biomarkers, potential therapeutic targets for various diseases such as cancer, diabetes, cardiovascular disease, and neuropathy. With further advances in technology and data analysis, LC–MS/MS-based metabolomics is expected to play an increasingly important role in future disease biology research.
References
(1) Han, J.; Lu, Q.; Chen, Y.; Yang, Y. Recent Metabolomics Analysis in Tumor Metabolism Reprogramming. Front. Mol. Biosci. 2021, 8, 1–16. DOI: 10.3389/fmolb.2021.763902
(2) Jin, Q.; Ma, R.C.W. Metabolomics in Diabetes and Diabetic Complications: Insights from Epidemiological Studies. Cells 2021, 10 (11), 2832. DOI: 10.3390/cells10112832
(3) McGarrah, R.W.; Crown, S.B.; Zhang, G.-F.; Shah, S.H.; Newgard, C.B. Cardiovascular Metabolomics. Circ. Res. 2018, 122 (9), 1238–1258. DOI: 10.1161/CIRCRESAHA.117.311002
(4) Pradhan, S.S.; Thota, S.M.; Rajaratnam, S.; et al. Integrated Multi-Omic Analysis of Huntington Disease Identifies Pathways that Modulate Protein Aggregation. Dis. Model Mech. 2022, 15 (10), dmm049492. DOI: 10.1242/dmm.049492
(5) Pradhan, S.S.; Rao, K.R.; Manjunath, M.; et al. Vitamin B6, B12 and Folate Modulate Deregulated Pathways and Protein Aggregation in Yeast Model of Huntington Disease. 3Biotech. 2023, 13 (3), 96. DOI: 10.1007/s13205-023-03525-y
(6) Rajaratnam, S.; Soman, A.P.; Phalguna, K.S.; et al. Integrated Omic Analysis Delineates Pathways Modulating Toxic TDP-43 Protein Aggregates in Amyotrophic Lateral Sclerosis. Cells 2023, 12 (9), 1228. DOI: 10.3390/cells12091228
(7) Thota, S.M.; Chan, K.L.; Pradhan, S.S.; et al. Multimodal Imaging and Visually Evoked Potentials Reveal Key Structural and Functional Features that Distinguish Symptomatic from Pre-Symptomatic Huntington’s Disease Brain. Neurol. India. 2021, 69 (5), 1247–1258. DOI: 10.4103/0028-3886.329528
(8) Johnson, K.A.; Fox, N.C.; Sperling, R.A.; Klunk, W.E. Brain Imaging in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012, 2 (4), a006213. DOI: 10.1101/cshperspect.a006213
(9) Pulukool, S.K.; Bhagavatham, S.K.S.; Kannan, V.; et al. Elevated Dimethylarginine, ATP, Cytokines, Metabolic Remodeling Involving Tryptophan Metabolism and Potential Microglial Inflammation Characterize Primary Open Angle Glaucoma. Sci Rep. 2021, 11, 17903. DOI: 10.1038/s41598-021-97509-8
(10) Kao, Y.-C.; Ho, P.-C.; Tu, Y.-K.; Jou, I.-M.; Tsai, K.-J. Lipids and Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21 (4), 1505. DOI: 10.3390/ijms21041505
(11) King, A.M.; Mullin, L.G.; Wilson, I.D.; et al. Development of a Rapid Profiling Method for the Analysis of Polar Analytes in Urine Using HILIC–MS and Ion Mobility Enabled HILIC–MS. Metabolomics 2019, 15, 17. DOI: 10.1007/s11306-019-1474-9
(12) Sathe, G.; Na, C.H.; Renuse, S.; et al. Quantitative Proteomic Profiling of Cerebrospinal Fluid to Identify Candidate Biomarkers for Alzheimer’s Disease. Proteomics Clin. Appl. 2018, 13 (4) e1800105. DOI: 10.1002/prca.201800105
Rajesh Babu Dandamudi is a business application manager in Phenomenex and manages Phenomenex Applications Lab in India. He has more than 13 years of chromatography experience and is an expert in practical HPLC/UHPLC/LC–MS/GC–MS method development and troubleshooting. He has more than 30 research publications to his credit. In his current role, Rajesh supports customers across India in method development and troubleshooting and also delivers on-site seminars.
Venketesh Sivaramakrishnan is an associate professor at Sri Sathya Sai Institute of Higher Learning in Puttaparthi, India. His primary focus is on disease biology. His research group, in collaboration with clinicians, carries out integrative analysis of clinical and multi-omics data analysis—mechanistic studies using model systems—and translates their findings using animal models. He has authored or coauthored more than 45 publications and has more than 20 conference presentations to his credit.

HILIC Peptide Retention Times Predicted Using New Approach
October 29th 2024Manitoba Centre for Proteomics and Systems Biology scientists produced a new means of predicting peptide retention times for hydrophilic interaction liquid chromatography (HILIC) at acidic pH in formic-acid based eluents.